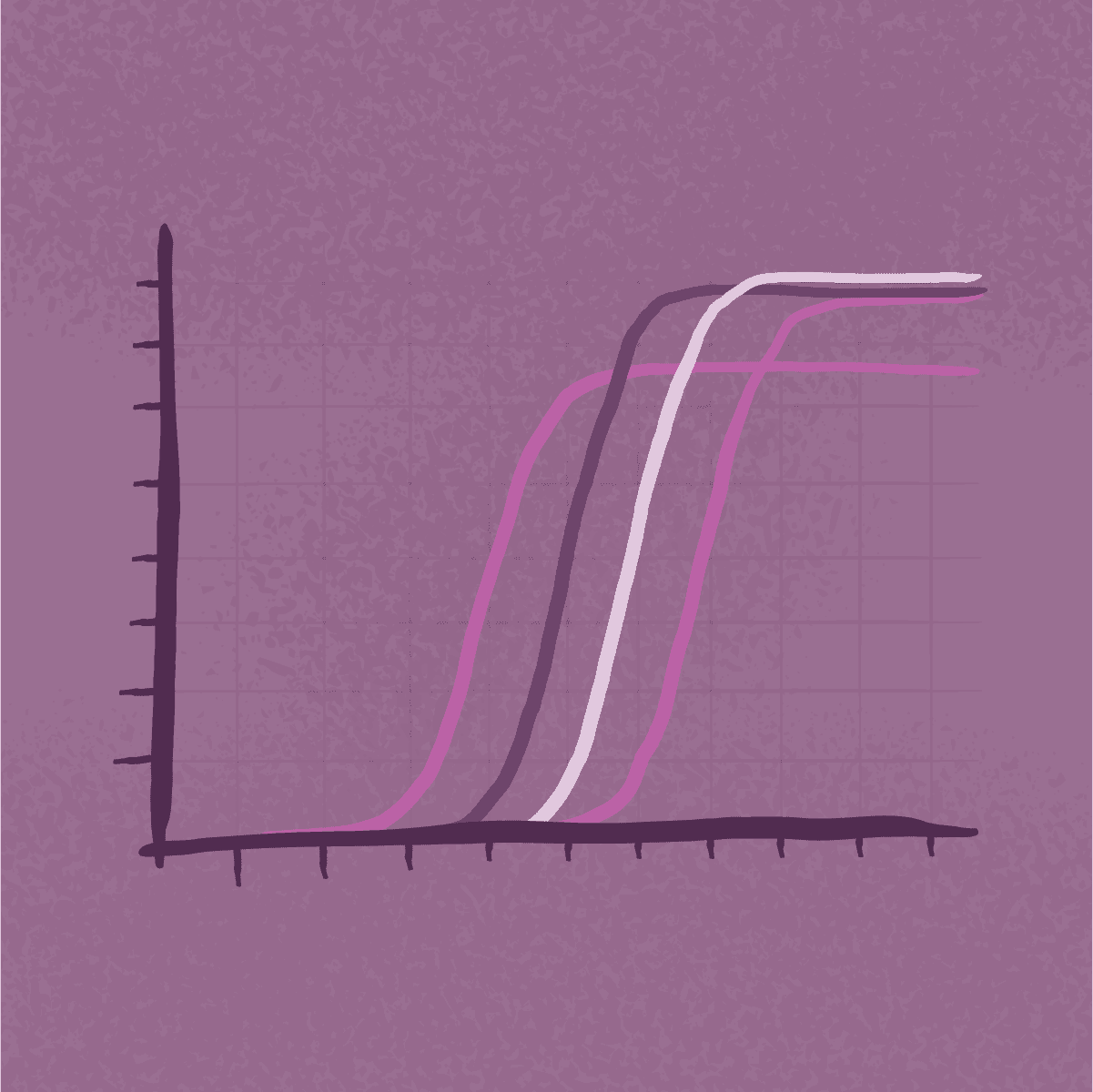Nucleic Acid Analysis
Nucleic acid analysis (genomics) involves isolation and characterization of DNA or RNA for use in applications such as genotyping, gene expression analysis, epigenetic analysis, microbiome studies and more. Experimental methods fundamental to nucleic acid analysis include nucleic acid extraction and clean up, DNA/RNA quantification, PCR, molecular cloning and sequencing.
Nucleic Acid Analysis Product Categories
Cloning and DNA Markers
Restriction enzymes, cloning vectors, molecular weight markers and competent cells.
Epigenetics
Products for DNA Methylation analysis, PCR and qPCR, reporter assays, HDAC assays, and protein purification systems.
Nucleic Acid Extraction
Adaptable extraction kits that generate high yields for many samples and throughputs.
PCR
High-quality enzymes and kits for all your amplification needs.
RNA Analysis
Nucleic acid analysis workflow products, as well as all-inclusive kits for RT-PCR.
Sequencing
Products and solutions for high-quality NGS input template or confirmatory testing.
Vectors
Basic cloning and subcloning vectors, reporter vectors and protein expression vectors.
An Introduction to Nucleic Acid Analysis
Purification, quantification and amplification of DNA and RNA are common methods used in many laboratory procedures across a wide variety of applications. Extraction of high-quality nucleic acid from cultured cells, animal or plant tissues, bacteria, body fluids and other sample types is a critical first step in most genomic analysis experiments. If the quality of the extracted DNA is poor, all downstream steps can be compromised.
During nucleic acid extraction and purification, samples progress though a common set of manipulations: cell lysis, clearing, nucleic acid binding, washing and elution of the purified DNA or RNA. The purity and yield of the extracted nucleic acid are key considerations for optimal performance in downstream applications such as PCR and sequencing.
Nucleic acid purification methods may be designed to isolate DNA or RNA for a wide range of samples or may be specific for an individual sample type. Most DNA purification kits can handle numerous sample types with minor protocol variations to ensure complete lysis of the starting material. Specialized purification kits may be needed for more challenging sample types that contain degraded DNA or a have low concentration of the target nucleic acid. Challenging sample types include formalin-fixed-paraffin-embedded (FFPE) tissue samples, plasma or serum containing circulating cell-free DNA, forensic samples, or any source where the sample quantity is limiting.
Once the nucleic acid is purified, most downstream assays require quantification of DNA or RNA before you begin. Nucleic acid quantification methods include measurement of fluorescent DNA or RNA binding dyes, measurement of absorbance at 260nm wavelength, and qPCR. Nucleic acid purity also provides a decision point to proceed with downstream assays or not based on 260/230 nm or 260/280 nm ratios.
Real-time quantitative PCR, or qPCR, is a standard nucleic acid analysis tool used for detection and quantification of gene expression, and for detection of specific DNA sequences, including mutations. Standard endpoint PCR and RT-PCR are used to detect and amplify specific DNA sequences for further manipulation, including analysis by sequencing.
Cloning methods, including restriction digestion, subcloning, PCR cloning and bacterial cell transformation are also still widely used in molecular biology labs. These classical methods for genetic manipulation of nucleic acids remain an important tool for genomic analysis.
The resources below provide in-depth information on nucleic acid analysis methods and link to products for each application.