HiBiT Protein Tagging System
Analyze and quantify proteins with high-sensitivity, tailored for your need.
HiBiT allows precise quantification and localization of proteins using simple, single-reagent bioluminescence methods. The adaptability and sensitivity provides new possibilities for studying protein function, degradation, secretion, and interactions.
What Is HiBiT Tagging?
HiBiT is a small, 11-amino-acid, epitope tag capable of producing a bioluminescent signal when bound to its complementation partner, LgBiT. HiBiT has unparalleled versatility and convenience with options for both simple and fast bioluminescent detection. The sensitive, high-affinity anti-HiBiT monoclonal antibody enables use in traditional immunoassays like Western blots and immunoprecipitation, making it adaptable to diverse workflows.
HiBiT technology offers an exceptional dynamic range, detecting low-abundance proteins at endogenous levels without requiring overexpression, thus preserving physiological relevance.
To obtain the HiBiT sequence, you must review the terms and conditions for use.

HiBiT Products
Protein Expression
Endogenous HiBiT Expression
HiBiT Overexpression
Protein Detection
Western Blot
Immunocytochemistry
Immunoprecipitation
Flow Cytometry
Protein Quantification
Intracellular Live-Cell Kinetic Quantification
- Nano-Glo® Endurazine™ and Vivazine® Live Cell Substrates
- LgBiT Expression Vector and Stable Cell Line
Cell-Surface or Secreted Protein Quantification
Total Protein Quantification
Functional & Cellular Assays
Target Cell Killing
Viral Neutralization
Autophagy
HiBiT Applications
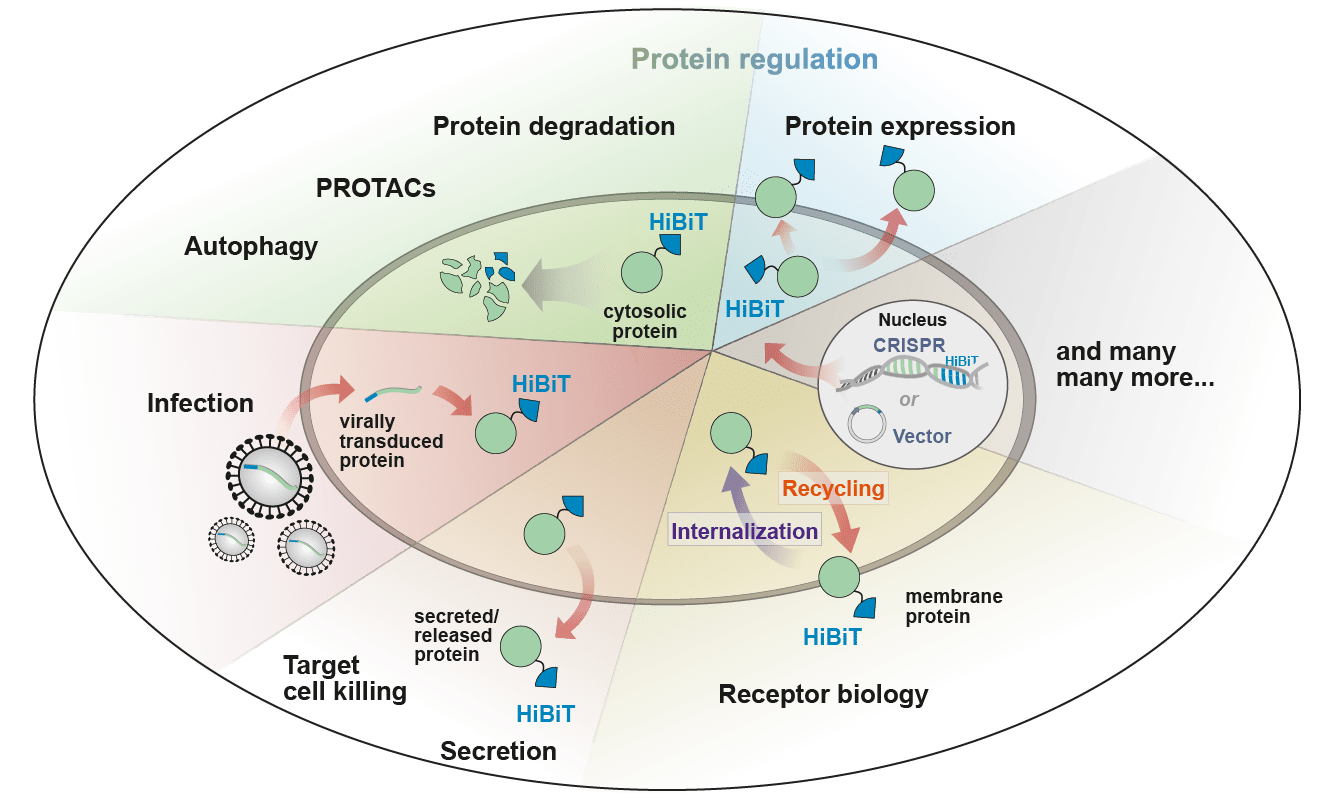
Receptor Biology
Read this article on how HiBiT tagging helped understand the trafficking of receptors involved in controlling appetite.
Intracellular Proteins
View this webinar on using HiBiT for high-throughput screening of RNA splicing modulators.
Cell-Based Potency Assessment
View this webinar about a HiBiT target-cell killing based approach for CAR-T potency assessment.
Targeted Protein Degradation
Read this article on using HiBiT for bioluminescence imaging to visualize target protein degradation.
Reduce Artifacts and Study Endogenously Expressed Proteins with CRISPR Technology
CRISPR insertion allows for monitoring of proteins under endogenous expression, removing artifacts observed from overexpression and more accurately modeling the biology. The small size of HiBiT makes insertion with CRISPR efficient, and the bioluminescence method enables sensitive, quantitative detection, even of proteins with low expression. The process removes the need for molecular cloning, reducing the time it takes to insert a knock-in tag from weeks to days.
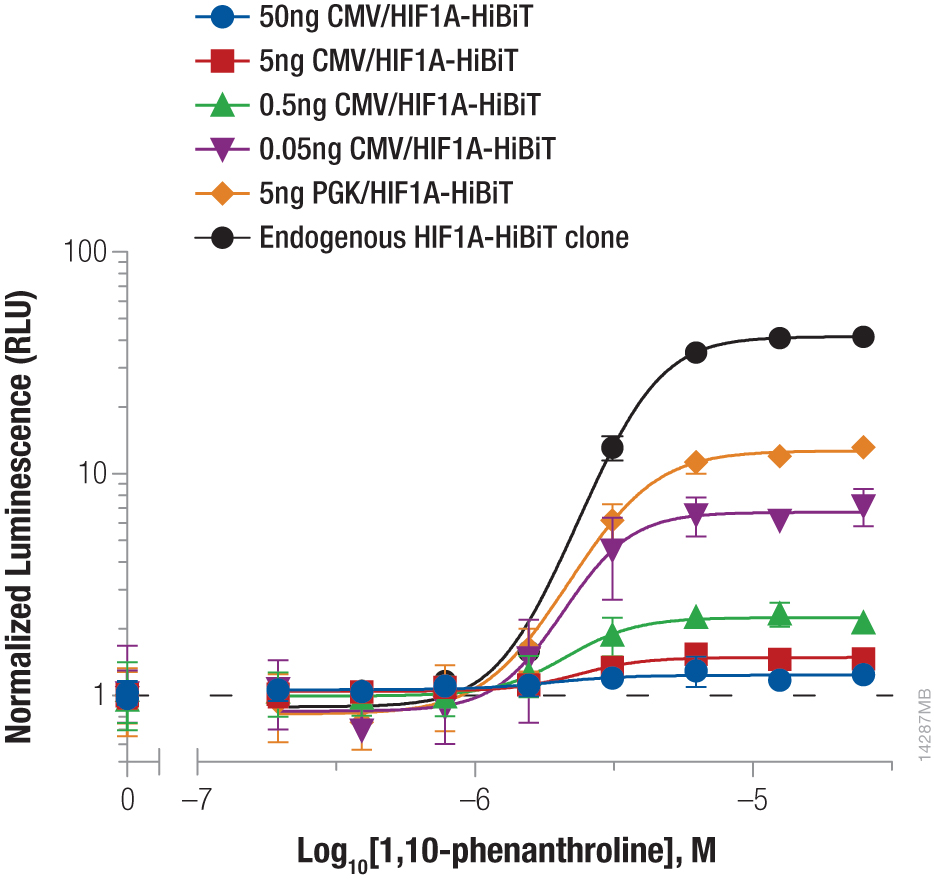
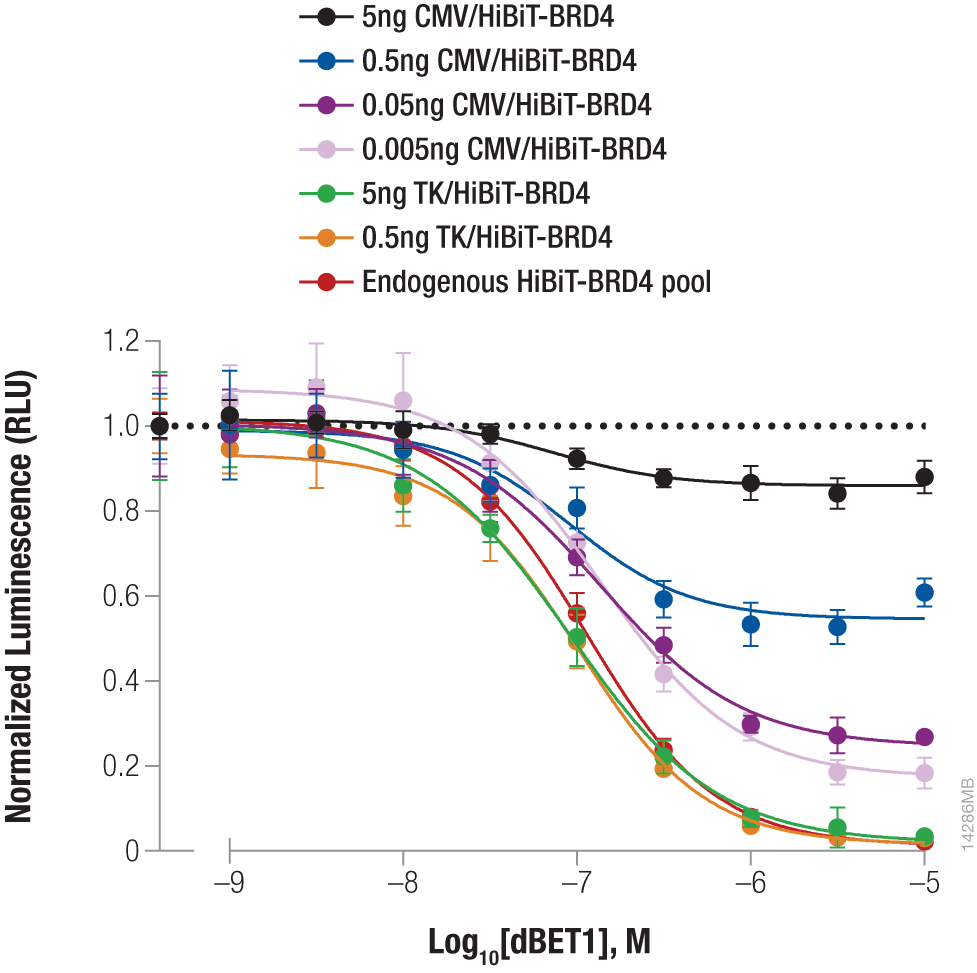
Left Panel: Stabilizing Hif1a-HiBiT by incubation with the hypoxia mimetic 1,10-phenanthroline. Right Panel: Targeted degradation of HiBiT-BRD4 by incubating with compound dBET1. See figure details in this article: Quantifying Protein Abundance at Endogenous Levels.