NanoBRET® Target Engagement
Establishing that a molecule engages with its intended target protein in cells is an important step in drug discovery and chemical probe development. NanoLuc® luciferase is the basis of the NanoBRET® Target Engagement assay platform that uses bioluminescence energy transfer to quantitatively measure target occupancy, compound affinity, residence time and permeability in live cells.

What is NanoBRET® Target Engagement?
Target engagement describes the interaction or binding of a chemical compound with its target protein in a living system. Target engagement assays are needed at multiple stages of drug development, including target validation, establishing structure activity relationships, and confirming mechanism of action (MOA).
Promega has developed a novel live-cell binding assay, called the NanoBRET® Target Engagement Assay. It uses an energy transfer technique known as bioluminescence resonance energy transfer (BRET) to measure binding between the target protein and small molecules in live cells. This energy transfer is based on two elements: Cellular expression of the target protein fused to NanoLuc® Luciferase and a cell-permeable fluorescent NanoBRET® tracer, which binds reversibly to the target protein.
An overview of NanoBRET® technology and its application for target engagement assays.
NanoBRET® TE Cellular Assays Can Measure Compound:
- Affinity and Occupancy: Quantitate how tightly a compound binds to a target and how much is bound.
- Selectivity: Quantitate intracellular binding of a compound to closely related proteins.
- Permeability: Calculate a compounds ability to get into cells.
- Residence Time: Assess how long a compound binds to target protein.
Getting Started Options:
- Optimized, ready-to-use NanoBRET® TE assays for key drug targets:
- Over 340 Kinase Targets
- CRBN and VHL E3 ligases
- 15 RAS Dimers, including hot spot mutants
- 5 RAF Dimers, including protomers and homodimers
- 9+ HDACs and BRDs
- 10+ PARPs
- NLRP3 Inflammasome
- Synthesize novel tracers from chemical probes or DEL hits using NanoBRET® 590 Dyes.
- Work with the Promega Services Team for compound profiling or new assay development.
How Does the NanoBRET® TE Assay Work?

Quantitate Affinity and Occupancy
Unlike other cellular target engagement assays, NanoBRET® TE assays are quantitative for intracellular compound affinity (or apparent Ki), not just potency. Quantitative affinity measurements are achieved by use of an appropriate NanoBRET® Tracer concentration, which is less than or equal to Tracer Kd. For each target specific NanoBRET® TE assay developed by Promega, the apparent intracellular NanoBRET® Tracer affinity and recommended Tracer concentration is provided.
NanoBRET® TE is also quantitative for fractional occupancy. By using the appropriate experimental controls, the BRET ratio can be converted to occupancy. Use of fractional occupancy allows quantifying compound selectivity across many similar targets, such as kinases.
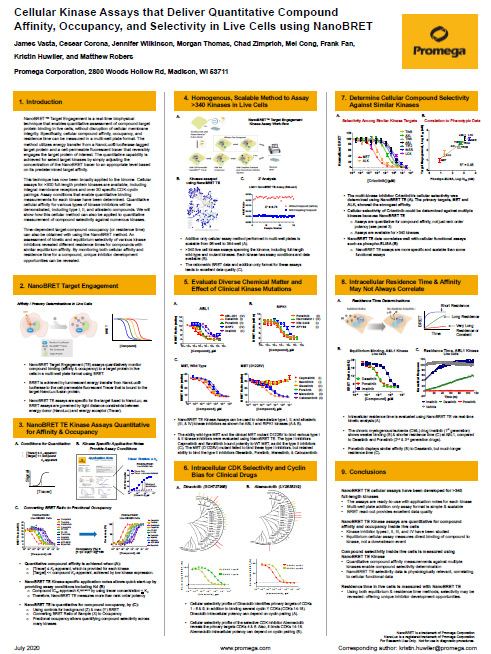
The poster Cellular Kinase Assays that Deliver Quantitative Compound Affinity, Occupancy, and Selectivity in Live Cells using NanoBRET provides details on how quantitation is achieved, as well as supporting data using NanoBRET® TE Kinase assays.
Target Engagement at Protein Complexes
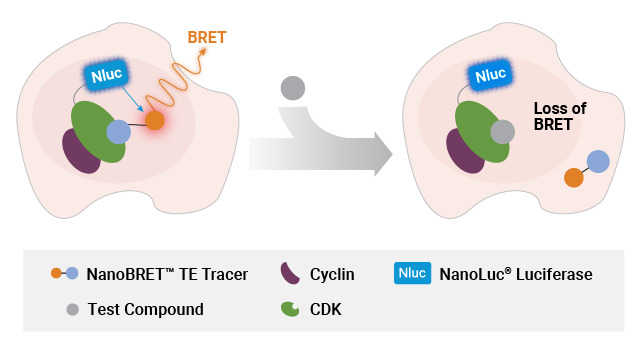
NanoBRET® TE assay can also quantify compound binding to target protein in a protein complex. In the first iteration of this application, target engagement at protein complexes is achieved by co-expressing in excess the binding partner with the target protein, which creates a biased readout specific for the target-partner protein pair. This approach is exemplified by the NanoBRET® TE assays for cyclin-dependent kinases (CDKs), which are typically assayed with the co-expression of a partner cyclin protein.
The use of NanoBRET® TE Assay to profile CDK inhibitors in live cells was described in the 2020 Nature Communications publication by Wells, C.I. et al.
In the latest iteration, the NanoBiT® System was incorporated into the NanoBRET® TE Assay to ensure that the BRET signal is conditional on the formation of a protein complex between the target protein and its binding partner. This approach was used in the NanoBRET® TE Assays for RAS proteins and RAF dimers, enabling the evaluation of target engagement at RAS or RAF proteins in multimeric complexes. Using the NanoBRET® TE Intracellular RAF Dimer Assays to profile RAF dimer inhibitors, we have observed differential compound affinity for CRAF depending on the CRAF dimers formed (see product page to learn more).
A.
B.
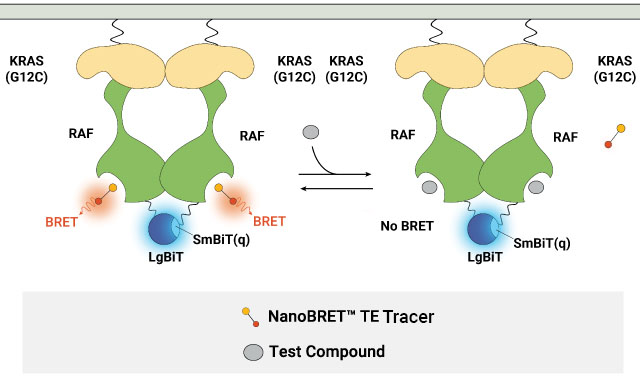
Measuring compound binding at protein complexes with NanoBRET® TE Assays. A) Principle of the NanoBRET® TE Assay for CDKs. Co-expression of a cyclin partner with the CDK-NanoLuc® fusion protein drives the formation of the CDK-cyclin complex. B) Schematic of the NanoBRET® TE Intracellular RAF Dimer Assays. RAF proteins are tagged with LgBiT or SmBiT(q) to ensure that the BRET signal is specific to the RAF dimer complex.
Cellular Selectivity Profiling
A key advantage of NanoBRET® TE is the ability to quantitatively measure the selectivity of a compound for multiple related proteins or mutants in live cells. This is possible because the NanoBRET® TE Assays can quantify the fractional occupancy and affinity of a compound across targets in live cells (see details above). An example of this is shown with the NanoBRET® TE Intracellular RAS Assay, where the cellular affinity of a RAS ligand is determined against 15 different KRAS and HRAS variants.
To perform selectivity profiling of a compound across a hundred or more targets, the ability of NanoBRET® TE to measure compound occupancy at all targets at a single concentration of compound is critical. An example of this selectivity analysis for kinase inhibitors is illustrated by the NanoBRET® TE K192 Kinase Selectivity System, which contains a panel of 192 kinases that broadly represent the kinome. Using this System, the cellular fractional occupancy at each kinase is determined for the compound of interest. We have often observed improved compound specificity in cells relative to biochemical approaches. The NanoBRET® TE CDK Selectivity Systems can be used for a CDK-focused selectivity profiling.
Want to have Promega perform the kinase selectivity profiling? Promega Tailored R&D Solutions team offers compound profiling services for many targets using the NanoBRET® TE Assays, including the K192 panel or a larger kinase panel.
A.

B.
NanoBRET® TE Cellular
Crizotinib, 16 hits
Biochemical
Crizotinib, 49 hits
Selectivity Profiling in Live Cells using NanoBRET® TE Assays. A) Example selectivity profiles for RAS inhibitor MRTX1133, which broadly engages KRAS and KRAS mutants but exhibits reduced affinity for HRAS variants. B) Comparison of target occupancy obtained with 1µM Crizotinib using the live-cell NanoBRET® TE K192 Kinase Selectivity System vs. a biochemical cell-free approach. The results showed an improved spectrum of specificity in cells.
Assess Intracellular Compound Availability
NanoBRET® TE assays can be used to assess the intracellular availability of compounds, which serves as a proxy for compound permeability. This is achieved by performing the NanoBRET® TE assays in two modes, a live-cell mode and a permeabilized-cell mode. In the live-cell mode, the assay measures the apparent cellular affinity, where the plasma membrane is intact and can impede the compound's access to its target. In the permeabilized-cell mode, the barrier posed by the plasma membrane is removed and the intrinsic affinity of the compound for target is measured. Affinity data from both modes is then used to calculate intracellular availability.
To facilitate the comparison of intracellular availability among compounds, a permeable control compound is included for normalization. This allows for the calculation of a parameter called the Availability Index (AI), which can be compared between compounds to assess relative intracellular availability, with compounds of higher intracellular availability having a smaller AI. Comparison of intracellular availability between compounds provides a means to prioritize compounds based on properties associated with permeability, which can play an important role in developing drug candidates of high molecular weights, such as PROTACs.
The poster NanoBRET as a Live Cell Method to Assess PROTAC Affinity & Intracellular Availability for E3 Ligases provides details on how intracellular availability can be assessed, using NanoBRET® TE E3 Ligase assays as an example.
Determine Residence Time
NanoBRET® Target Engagement Assays have the unique ability to assess compound residence time in live cells. This measurement involves using cells expressing the target-NanoLuc® fusion protein and equilibrating them with a near-saturating concentration of compound. Next, unbound compound is removed and cells are treated with a near-saturating concentration of tracer. The binding is followed kinetically such that compounds with slow dissociation kinetics from the target impede tracer binding, which slows production of the BRET signal. The real-time monitoring of binding kinetics and drug-target residence time in live cells is enabled by fast-binding NanoBRET® tracers and the long signal half-Life for NanoLuc® Luciferase.
See how researchers at AstraZeneca used the NanoBRET® TE assay to determine compound residence time and uncovered kinetic elements in target selectivity.
A.

B.

C.

Measuring compound residence time with NanoBRET® technology. A) Overview of the workflow. B) Phenotypic potency of HDAC inhibitors. Treatment with HDAC inhibitors for 48 hours results in antiproliferative effects in HeLa cells, as measured by intracellular ATP levels. C) Residence time analysis reveals a remarkably slow dissociation rate for FK228 from HDAC1, providing an explanation for the prolonged phenotypic effect observed with FK228.
Design Your Own Target Engagement Assay
NanoBRET® Target Engagement technology is applicable to multiple classes of target proteins, including kinases, HDACs, bromodomains and GPCRs. If there is no NanoBRET® TE Assay for your target of interest, you can create one! There are three stages to developing a NanoBRET® TE Assay:
- Synthesize fluorescent tracer candidates that can bind the target of interest. The NanoBRET® 590 dyes are optimal for generating your novel NanoBRET® tracers.
- Construct vectors that express a fusion of your target protein with NanoLuc® luciferase. You have the option of using either the Flexi® Vector System-compatible vectors or vectors with multiple cloning sites.
- Evaluate the performance of tracer candidates in NanoBRET® TE assay. If your target of interest is an intracellular protein, you will need the Intracellular TE Nano-Glo® Substrate/Inhibitor. The components provided in this product will ensure that the BRET signal measured is the result of intracellular interactions.
To learn more about Do-It-Yourself NanoBRET® TE assay, visit the product page for the NanoBRET® 590 dyes.

Additional Resources
Quantifying Residence Time and Target Binding
Learn how a novel BRET assay is able to assess cellular target engagement and residence time in this webinar.
Measuring Kinase Target Engagement in Live Cells
This webinar describes how to examine target engagement for kinases.
The Surprising Landscape of CDK Inhibitor Selectivity in Live Cells
This blog post describes a study to repurpose some kinase inhibitors as selective chemical probes for lesser-studied CDK family members.