DNA Purification
For ordering information on the products discussed here, please visit our Nucleic Acid Extraction product pages.
Introduction
In today’s world of DNA analysis by multiplex and real-time PCR, the importance of high-quality, purified DNA cannot be underestimated. Finding a suitable DNA isolation system to satisfy your downstream application needs is vital for the successful completion of experiments.
This DNA purification guide addresses general information on the basics of DNA extraction, plasmid preparation and DNA quantitation, as well as how optimized purification techniques can help increase your productivity, so you spend less time purifying DNA and more time developing experiments and analyzing data.
In addition, this guide covers the wide variety of Promega products available for genomic, plasmid and fragment/PCR product purification. Along with the discussion of Promega’s DNA extraction systems, we also consider the issues of scalability, purity, yield and the effects they have on downstream applications, to assist in finding the best system for your needs.
DNA Purification Basics
Basic Isolation Procedure
There are five basic steps of DNA extraction that are consistent across all the possible DNA purification chemistries: 1) disruption of the cellular structure to create a lysate, 2) separation of the soluble DNA from cell debris and other insoluble material, 3) binding the DNA of interest to a purification matrix, 4) washing proteins and other contaminants away from the matrix and 5) elution of the DNA.
1. Creation of Lysate
The first step in any nucleic acid purification reaction is releasing the DNA/RNA into solution. The goal of lysis is to rapidly and completely disrupt cells in a sample to release nucleic acid into the lysate. There are four general techniques for lysing materials: physical methods, enzymatic methods, chemical methods and combinations of the three.
Physical Methods
Physical methods typically involve some type of sample grinding or crushing to disrupt the cell walls or tough tissue. A common method of physical disruption is freezing and grinding samples with a mortar and pestle under liquid nitrogen to provide a powdered material that is then exposed to chemical or enzymatic lysis conditions. Grinders can be simple manual devices or automated, capable of disruption of multiple 96-well plates. Physical methods are often used with more structured input materials, such as tissues or plants. Other devices use bead beating or shaking in the presence of metallic or ceramic beads to disrupt cells or tissues, or sonication to disrupt tissues and lyse cells.
Chemical Methods
Chemical methods can be used alone with easy-to-lyse materials, such as tissue culture cells or in combination with other methods. Cellular disruption is accomplished with a variety of agents that disrupt cell membranes and denatures proteins. Chemicals commonly used include detergents (e.g., SDS) and chaotropes (e.g., guanidine salts and alkaline solutions).
Enzymatic Methods
Enzymatic methods are often used with more structured starting materials in combination with other methods with tissues, plant materials, bacteria and yeast. The enzymes utilized help to disrupt tissues and tough cell walls. Depending on the starting material, typical enzymatic treatments can include: lysozyme, zymolase and liticase, proteinase K, collagenase and lipase, among others. Enzymatic treatments can be amenable to high throughput processing, but may have a higher per sample cost compared to other disruption methods.
In many protocols, a combination of chemical disruption and another is often used since chemical disruption of cells rapidly inactivates proteins, including nucleases.
2. Clearing of Lysate
Depending on the starting material, cellular lysates may need to have cellular debris removed prior to nucleic acid purification to reduce the carryover of unwanted materials (proteins, lipids and saccharides from cellular structures) into the purification reaction, which can clog membranes or interfere with downstream applications. Usually clearing is accomplished by centrifugation, filtration or bead-based methods.
Centrifugation can require more hands-on time, but it is able to address large amounts of debris. Filtering can be a rapid method, but samples with a large amount of debris can clog the filter. Bead-based clearing, like the method used with Promega particle-based plasmid prep kits, can be used in automated protocols, but can be overwhelmed with biomass. Once a cleared lysate is generated, the DNA can then be purified by many different chemistries, such as silica, ion exchange, cellulose or precipitation-based methods.
3. Binding to the Purification Matrix
Regardless of the method used to create a cleared lysate, the DNA of interest can be isolated using a variety of different methods. Promega offers genomic DNA isolation systems based on sample lysis by detergents, and purification by binding to matrices (silica, cellulose and ion exchange), which is where interest has primarily been focused in recent years.
Each of these chemistries can influence the efficiency and purity of the isolation, and each have a characteristic binding capacity. Bind capacity is an indication of how much nucleic acid an isolation chemistry can bind before it reaches the capacity of the system and no longer isolates more of that nucleic acid. We can build design features into these chemistries by manipulating the binding conditions to enrich for different categories of nucleic acid (e.g., chemistries that selectively bind RNA versus DNA or large versus small fragments).
Solution-Based Chemistry
This type of chemistry does not rely on a binding matrix, but rather on alcohol precipitation. Following the creation of lysate, the cell debris and proteins are precipitated using a high-concentration salt solution. The high concentration of salt causes the proteins to fall out of solution, and then centrifugation separates the soluble nucleic acid from the cell debris and precipitated protein (1).
The DNA is then precipitated by adding isopropanol to the high-concentration salt solution. This forces the large genomic DNA molecules out of solution, while the smaller RNA fragments remain soluble. The insoluble DNA is then pelleted and separated from salt, isopropanol and RNA fragments via centrifugation.
Additional washing of the pellet with ethanol removes the remaining salt and enhances evaporation. Lastly, the DNA pellet is resuspended in an aqueous buffer like Tris-EDTA or nuclease-free water and, once dissolved, is ready for use in downstream applications.
Silica-Binding Chemistry
The technology for these genomic DNA purification systems is based on binding of the DNA to silica under high-salt conditions (2–4). The key to isolating any nucleic acid with silica is the presence of a chaotropic salt like guanidine hydrochloride. Chaotropic salts present in high quantities are able to disrupt cells, deactivate nucleases and allow nucleic acid to bind to silica.
Once the genomic DNA is bound to the silica membrane, the nucleic acid is washed with a salt/ethanol solution. These washes remove contaminating proteins, lipopolysaccharides and small RNAs to increase purity while keeping the DNA bound to the silica membrane column. Once the washes are finished, the genomic DNA is eluted under low-salt conditions using either nuclease-free water or TE buffer.
Binding to silica is not DNA specific, so if pure DNA is required, there is also the option to add ribonuclease (RNase A) to the elution buffer. RNA may be may be copurified with gDNA, and the addition of RNase to the elution buffer ensures the removal of the vast majority of contaminating RNA.
This chemistry can be adapted to either paramagnetic particles (PMPs), like Promega silica-coated MagneSil® PMPs, or silica membrane column-based formats. While both methods generally represent a good balance of yield and purity, the silica membrane column format is more convenient. For automated purification, either the 96-well silica membrane plates or the MagneSil® PMPs are easily adapted to a variety of robotic platforms.
In order to process the DNA samples, the MagneSil® PMPs require a strong magnet for particle capture, rather than centrifugation or vacuum filtration. The MagneSil® PMPs are considered a “mobile solid phase” with binding of nucleic acids occurring in solution. Particles can also be completely resuspended during the wash steps of a purification protocol, thus enhancing the removal of contaminants. See Figure 1 for images of a silica membrane column and the MagneSil® PMPs.
Cellulose-Binding Chemistry
More recently, Promega has commercialized DNA isolation methods that use a cellulose-based matrix. Nucleic acid binds to cellulose in the presence of high salt and alcohols. Generally speaking, the binding capacity of cellulose-based methods is very high. Conditions can be adjusted to preferentially bind different species and sizes of nucleic acid. As a result of the combination of binding capacity and relatively small elution volume, we can get high concentration eluates for nucleic acids.
Ion Exchange Chemistry
Ion exchange chemistry is based on the interaction that occurs between positively-charged particles and the negatively-charged phosphates that are present in DNA. The DNA binds under low salt conditions, and contaminating proteins and RNA can then be washed away with higher salt solutions. The DNA is eluted under high salt conditions, and then recovered by ethanol precipitation.
4. Washing
Wash buffers generally contain alcohols and can be used to remove proteins, salts and other contaminants from the sample or the upstream binding buffers. Alcohols additionally help associate nucleic acid with the matrix.
5. Elution
DNA is soluble in low-ionic-strength solution such as TE buffer or nuclease-free water. When such an aqueous buffer is applied to a silica membrane, the DNA is released from the silica, and the eluate is collected. The purified, high-quality DNA is then ready to use in a wide variety of demanding downstream applications, such as multiplex PCR, coupled in vitro transcription/translation systems, transfection and sequencing reactions.
When selecting your elution buffer, it is important to consider the requirements of your desired downstream processes. Eluting and storing the DNA in TE buffer, for example, is helpful as long as the EDTA does not impact your chosen downstream applications. EDTA chelates, or binds, magnesium present in the purified DNA and can help inhibit possible contaminating nuclease activity. If EDTA is a concern, we recommend storing DNA in a buffered solution, as the acidic nature of DNA can lead to autohydrolysis. Alternatively, you can use TE-4 buffer, which is 10mm Tris-HCl, 0.1mm EDTA (pH 8.0).
Wizard® SV Genomic DNA Purification System
A fast, simple, silica membrane-based technique for preparing genomic DNA from cultured cells and tissue.
ReliaPrep™ Blood gDNA Miniprep System
A cellulose column-based, ready-to-use system that obtains intact genomic DNA without using ethanol washes or precipitations.
Genomic DNA Isolation
Yield, purity and integrity are essential to performance in downstream applications such as PCR and sequencing. Optimization of extraction methodologies is key for success with challenging sample types and demanding downstream applications. The purified target DNA should be free of contaminants, including proteins, other cellular components and undesired nucleic acids.
Specialized, sample-type specific purification kits may be needed for more complex and challenging samples that contain degraded DNA or have low concentrations of DNA. Challenging sample types include FFPE tissue, plasma or serum containing cell-free DNA, forensic samples or any source where the sample quantity is limiting.
Promega was one of the first companies to provide kits for the purification of DNA, as well as plasmids, with over 30 years of experience in nucleic acid extraction. We offer a wide range of genomic DNA extraction kits suitable for a variety of sample types and throughput needs, producing high yields and high-quality DNA for use in your downstream applications. Our products cover a variety of throughput options and processing methods suitable to your specific needs—from manual single-preps to small benchtop or large-scale automated systems.
Utilizing spin, vacuum or magnetic-based methods, our manual single-prep solutions are best for processing less than 24 samples at a time. If you are looking for an automated solution, our cartridge-based kits for use with Maxwell® Instruments can process up to 48 samples in the same run. We also offer fully automated high-throughput extraction options utilizing plate-based processing methods, fully compatible with liquid handling platforms.
Although techniques like Southern blotting, which require microgram amounts of DNA, are still performed in molecular biology laboratories, most assessments of chromosomal DNA is done by PCR-based technologies. These include monoplex or multiplex PCR, SNP arrays, analysis and real-time PCR, ddPCR and next-generation sequencing (NGS). These latter techniques use nanogram amounts of DNA per reaction. Regardless of the system chosen, Promega genomic DNA purification kits provide the required yields of high-quality DNA with minimal contaminants.
Manual Purification Systems
Solution-Based Systems
Promega offers genomic DNA isolation systems based on sample lysis by detergents and purification by various methods. These include both membrane-based systems (e.g., the single-column Wizard® SV Genomic DNA Purification System (Cat.# A2360, A2361) or the high-throughput, 96-well Wizard® SV 96 Genomic DNA Purification System (Cat.# A2370, A2371) and easily automated paramagnetic silica systems. All of these systems purify genomic DNA that is amenable for use in many downstream applications.
The Wizard® Genomic DNA Purification Kit (Cat.# A1120, A1125, A1620) is both a versatile and scalable system for isolating genomic DNA using a precipitation-based method. With this system alone, chromosomal DNA can be isolated from whole blood (5), plant leaf (6), Gram-positive (7) and Gram-negative bacteria (8), mouse tail (9) and yeast (10). Additional sample types like fungus (11), infected frog tissues embedded in paraffin (12), saliva (13) and flour beetles (14) have also been used successfully.
Not only is this genomic purification system successful with many sample types, it is also easily scaled for the quantity of starting material by adjusting reagent volumes to accommodate your needs.
Column-Based Systems
Traditional Column-Based Systems
For single-column isolation, the Wizard® SV Genomic DNA Purification System provides a fast, simple technique for the preparation of purified and intact DNA from mouse tails, tissues and cultured cells in as little as 20 minutes, depending on the number of samples processed (up to 24 by centrifugation, depending on the rotor size, or up to 20 by vacuum). A vacuum manifold or a microcentrifuge is used for sample processing. With some modifications, whole blood can also be used with this isolation system (15). This is a silica membrane-based system, meaning there are limitations to the amount of material that can be loaded onto a single SV column; up to 20mg of tissue (mouse tail or animal tissue) or between 1 × 104 and 5 × 106 tissue culture cells can be processed per purification. With more sample, the prepared lysate may need to be split among two or more columns to avoid clogging.
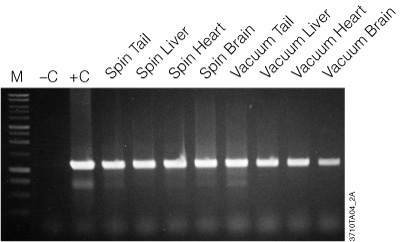
Figure 2. Amplification of genomic DNA isolated from various tissue sources using the Wizard® SV Genomic DNA Purification System. One microliter of purified genomic DNA was amplified using PCR Master Mix (Cat.# M7502) and mouse-specific IL-1β primers (1.2kb product). Reactions with Mouse Genomic DNA (Cat.# G3091; +C) and without DNA (–C) were performed as positive and negative controls, respectively. Thermal cycling conditions were: one cycle of 3 minutes at 95°C; followed by 30 cycles of: 95°C for 30 seconds, 60°C for 1 minute, 70°C for 1 minute and 30 seconds; final extension at 70°C for 7 minutes; 4°C soak. All lanes contained 10µl of reaction product separated on a 1% agarose gel. PCR products were visualized by ethidium bromide staining. “Spin” and “Vacuum” designations indicate the protocol used for genomic DNA isolation.
The genomic DNA isolated with the Wizard® SV Genomic DNA Purification System is of high quality and performs well in agarose gel analysis, restriction enzyme digestions and PCR analysis as seen in Figure 2. Table 1 provides typical yields of genomic DNA purified from a variety of sources.
Table 1. Typical Genomic DNA Yield From Various Tissues using the Wizard® SV Genomic DNA Purification System.
| Sample | Amount | Average Yield |
|---|---|---|
| Tail Clipping | 20mg | 20µg |
| Liver | 20mg | 15µg |
| Heart | 20mg | 10µg |
| Brain | 20mg | 6µg |
| CHO cells | 1 × 106 | 5µg |
| NIH/3T3 cells | 1 × 106 | 9µg |
| 293 cells | 1 × 106 | 8µg |
Researchers have used this simple and rapid system for many additional sample types and applications including mosquitoes (16), mammary stem cells (17), Bacillus subtilis (18), Escherichia coli (19), the larval form of the Schistosoma mansoni parasite (20) and viral DNA from Kaposi’s sarcoma herpes virus-infected BC3 cells (21).
For high-throughput, 96-well isolation, the Wizard® SV 96 Genomic DNA Purification System is available. Amplifiable genomic DNA can be isolated from up to 5 × 106 cells, 20mg of tissue or up to 1.2cm of a mouse tail tip without centrifugation of the lysate prior to purification.
This multiwell system requires a vacuum manifold (Vac-Man® 96 Vacuum Manifold, Cat.# A2291) and a vacuum pump capable of generating 15–20 inches of mercury or the equivalent. Genomic DNA was isolated from three different source types then used in a monoplex PCR and run on an agarose gel as shown in Figure 3. Figure 4 compares the yield from the three Wizard® SV Genomic DNA purification methods (96-well plate, vacuum and centrifugation).

Figure 3. Agarose gel electrophoresis of PCR products amplified from 1µl of mouse tail, CHO cells and tomato leaf sample genomic DNA isolated using the Wizard® SV 96 Genomic DNA Purification System. A total of 10µl of PCR product is visualized on a 1.5% agarose gel stained with ethidium bromide. Panel A. IL-1β (1.2kb) amplified from mouse tail. Panel B. β-actin (250bp) amplified from CHO cells. Panel C. Chloroplast DNA (600bp) amplified from tomato leaf. Lane M, 1kb DNA Ladder (Cat.# G5711).
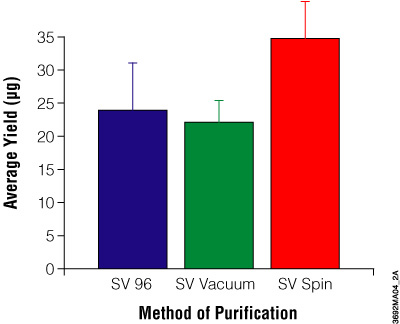
Figure 4. Comparison of DNA yields using the Wizard® SV and SV 96 Genomic DNA Purification Systems. Average yield of genomic DNA in micrograms purified from 20mg mouse tail clippings. The average A260/A280 ratios are: SV 96, 1.7 ± 0.08; SV vacuum method, 1.7 ± 0.14; SV spin method, 1.7 ± 0.14.
High-Performance Column-Based Systems
We offer two different ReliaPrep™ gDNA Miniprep Systems that purify genomic DNA using a cellulose column-based method: ReliaPrep™ Blood gDNA Miniprep System (Cat.# A5081, A5082) and ReliaPrep™ gDNA Tissue Miniprep System (Cat.# A2051, A2052). Both are ready-to-use systems that obtain intact genomic DNA without using ethanol washes or precipitations. The ReliaPrep™ Blood gDNA Miniprep System processes 200μl of blood or body fluid, either fresh or frozen, in less than 40 minutes. Yields from blood are typically 4–10μg, depending on the white blood cell count. Up to 25mg of tissue, a buccal (cheek) swab or a 1cm mouse tail can be processed with the ReliaPrep™ gDNA Tissue Miniprep System and the eluted DNA recovered in 30 minutes or less. The purified DNA can be eluted in as little as 50µl and is suitable for use in downstream applications such as RT-qPCR.
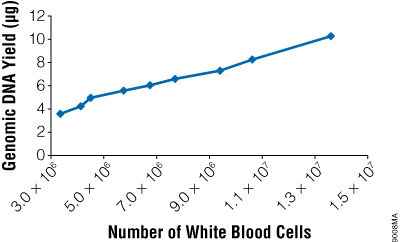
Figure 5. The yield of genomic DNA from the ReliaPrep™ Blood gDNA Miniprep System varies with white blood cell count. Whole blood was obtained from several individuals, and white cell counts were determined using a hemocytometer. Two hundred microliters of blood was used for genomic DNA purification (n = 3 or 4), and the amount of isolated gDNA was quantitated by absorbance spectroscopy.

Figure 6. Comparison of elution volume with concentration, yield and purity. Aliquots of blood (200μl) were processed using the ReliaPrep™ Blood gDNA Miniprep System (n = 4) and eluted with 30–200μl of Nuclease-Free Water. Concentration (Panel A), total yield (Panel B) and purity (Panel C) were assessed using absorbance spectroscopy. Yield decreased slightly with decreases in elution volume, while concentration increased. Purity as measured by optical density ratios remained constant.
Automated Systems for DNA Purification
As laboratories try to improve productivity for research, diagnostics and applied testing, the need has increased for easy-to-use, low- to moderate-throughput automation of purification processes. Automation eliminates the hands-on time and labor of manual purification, giving you more time and energy to focus on your research.
Cartridge-Based Systems
Traditionally, automation refers to the use of large, specialized and costly equipment that requires extensive training to operate and maintain. Promega has developed the Maxwell® Systems, which provide flexible, reliable, compact and easy-to-use alternatives to traditional automated systems.
The Maxwell® Systems are designed for efficient, automated purification from a wide range of sample types (see Table 2). Maxwell® Instruments are supplied with preprogrammed automated purification methods, and can process up to 48 samples in as little as 30–40 minutes (depending on instrument, sample type and method). The purified concentrated DNA or RNA are high quality and high yield, making them compatible with many common downstream applications, including qPCR, ddPCR, genotyping, sequencing and NGS.

Figure 7. The Maxwell® RSC (left) and Maxwell® RSC 48 (right).
Table 2. DNA yield from various sample types after purification using the Maxwell® RSC Instrument and DNA Purification Kits.
| Sample Type | Sample Size | Yield |
|---|---|---|
| Whole blood | 500µl whole blood | 60µg |
| Tissue |
Up to 50mg of liver tissue |
60µg 50µg |
| Cells | Up to 8 × 106 of cultured cells | 18µg |
| FFPE | Up to 2.0mm3 of mouse colon | 1.2µg |
| Bacteria | Up to 2 × 109 cells | 9.4µg |
| Plant | Up to 20mg of corn leaf | 100µg |
| Saliva | Up to 1ml of saliva | 1.11µg |
| Buccal Swab | 1 swab | 0.03µg |
| Serum-Plasma | Up to 1ml | 0.01µg |
| Buffy Coat | Up to 250µl | 200µg |
Maxwell® Kits offer predispensed reagent cartridges for purification of genomic DNA, RNA and Total Nucleic Acid. Application and sample type-focused kits make the Maxwell® Instruments a versatile extraction instrument for laboratories that may work with one or all of these different applications.
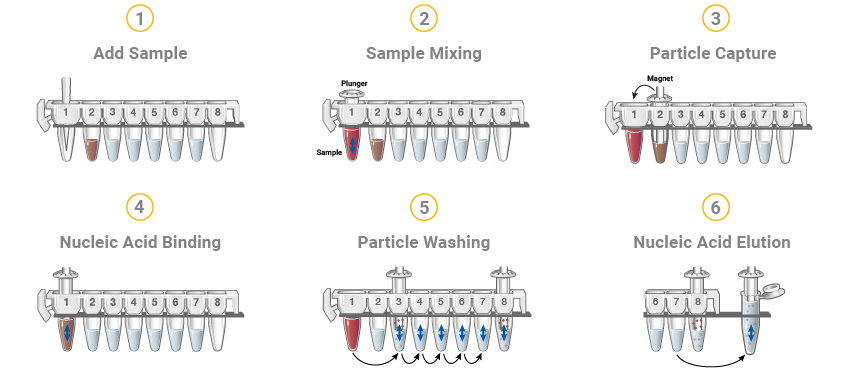
Figure 8. The Maxwell® RSC DNA or RNA extraction methods start with cartridges prefilled with purification reagents and paramagnetic particles, ready for your samples. After sample addition, the Maxwell® RSC moves the paramagnetic particles and associated nucleic acids through multiple steps ultimately yielding highly pure RNA or DNA in 30–100µl.
The Maxwell® Systems purify samples using paramagnetic particles (PMPs), which provide a mobile solid phase that optimizes sample capture, washing and elution of the nucleic acid. The Maxwell® Instruments are magnetic-particle-handling instruments that efficiently bind nucleic acids to the paramagnetic particle in the first well of a prefilled cartridge. The samples are processed through a series of washes before the nucleic acid is eluted. The systematic magnetic particle-based methodology used by the Maxwell® Instruments avoid common problems associated with automated liquid handler-based purification systems, such as clogged tips or partial reagent transfers, which can result in suboptimal purification processing.
The benchtop-compact Maxwell® Instruments are easy to set up and require no special training for use. Optimized automated methods are preloaded, the prefilled reagent cartridges are snapped into place, your sample is added and you select "Start" to begin the appropriate method. A full list of nucleic acid extraction kits is available here.
Several Maxwell® Instrument reagent kits are available and allow optimal extraction from a variety of sample types, including blood, serum and plasma, formalin-fixed, paraffin-embedded (FFPE) tissue, bacteria, plant, food and animal tissue.
Maxwell® HT Systems allow purification of DNA or RNA at scale on any laboratory liquid handler in 24- or 96-well SLAS format. Maxwell® purification chemistries use novel magnetic particle-based solutions that naturally decrease contamination carryover.
In addition to trusted chemistry, you’ll gain expert support to get started with automation or optimize your current HT workflow. Our team of automation experts can offer assistance with most of the leading laboratory automation providers in the world and help you develop and implement an automated nucleic acid purification solution customized to the needs of your laboratory.
Genomic DNA Extraction Kits
Looking for extraction options by sample scale or type? Explore our DNA extraction portfolio to discover the right solution for your purification needs.
Scalable Automation Solutions
The Maxwell® RSC Instruments provide a compact, automated nucleic acid purification platform that processes up to 16 (Maxwell® RSC) or up to 48 (Maxwell® RSC 48) samples simultaneously.
High-throughput Purification Chemistries and Automation Support
Maxwell® HT chemistries allow automation of nucleic acid purification on liquid handlers. Our team of automation experts offer assistance to help develop and implement an automated nucleic acid purification solution customized to the needs of your laboratory.
High-Throughput Systems for Genomic DNA Isolation
Promega offers several automated high-throughput options to isolate genomic DNA isolation from blood samples. Some laboratories, such as biobanks, have a desire to isolate DNA from large amounts of starting material (e.g., 10ml of blood). The ReliaPrep™ Large Volume HT gDNA Isolation System (Cat.# A2751) provides an effective means for isolation of genomic DNA derived from blood fractions derived from 2.5–10ml samples of whole blood. This chemistry can be automated onto liquid handlers by using a Promega HSM device, which enable processing of purification reactions in 50ml conical tubes.
Liquid level sensing and instrument operating software scale the chemistry to sample input volume for each individual sample, reducing reagent waste and expense. The automated system can also process sample in 14ml tubes using the Low Volume Adapter XAT1020 (LVA and Methods) which enables processing samples from 0.25–3ml.
There are no tedious centrifugation steps or hazardous chemicals, which are inherently handling workstation, offering walkaway purification of genomic DNA from whole blood, regardless of sample storage or shipping conditions.
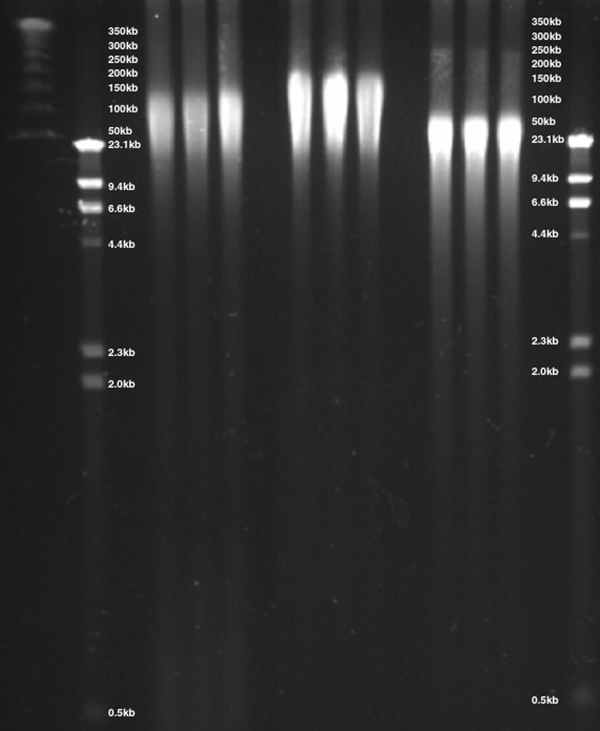
Figure 9. DNA was isolated from whole blood via three methods, separated by CHEF gel electrophoresis and visualized by ethidium bromide staining. DNA isolated using the ReliaPrep™ Large Volume HT gDNA Isolation System provided DNA with a size range of 20–125kb precipitation-based purification isolated DNA with a size range of 20–200kb while column-based methods demonstrated gDNA with a size of 20–75kb.
There is an option for low-throughput isolation of gDNA from up to 32 samples at one time when the Heater Shaker Magnet Instrument (HSM 2.0; Cat.# A2715) is used on a bench versus integrated on a liquid handler where the user dispenses and aspirates reagents from the samples as directed by the software on a computer screen. The preprogrammed methods control the heating, shaking, magnetization and timing of the steps required for the semi-automated purification.
In addition to whole blood, a variety of other sample types can also be processed, including stabilized saliva, buccal wash samples, blood fractions, buffy coats, red cell pellets and all cell pellets. For fully automated purification, the HSM 2.0 Instrument can be integrated with a robotic liquid-handling workstation.
Automating reagents onto instrumentation requires a carefully planned and executed approach. Collaborating with Promega gives you access to scientists who have designed automated purification for hundreds of labs, across a wide range of sample types.
Automating reagents onto instrumentation requires a carefully planned and executed approach. Collaborating with Promega gives you access to scientists who have designed automated purification for hundreds of labs, across a wide range of sample types.


Figure 10. Automated DNA yields for blood fractions. DNA yield is linear with respect to original volumes of blood. Panel A. DNA yields as determined by NanoDrop spectrophotometer. Panel B. DNA yields as determined using the QuantiFluor™ dsDNA System. All samples were prepared from a single donor. Manual samples were processed using the Wizard® Genomic DNA Purification Kit. Each point is the mean of n=4 values with error bars of 1 standard deviation.
Custom HT Nucleic Acid Purification
Implementing automated nucleic acid purification technologies onto your high-throughput workflow can be challenging and time-consuming. Our Field Support Scientists can provide the support you need to get started.
Selected DNA Purification Kits by Sample Type
Learn more about some of our specialized kits below, and explore the breadth of our portfolio and compare our DNA extraction kits with the help of our product comparison page to discover the right solution for your DNA purification needs.
Fixed-Tissue Genomic DNA Isolation
The MagneSil® Genomic, Fixed-Tissue System (Cat.# MD1490), provides a fast, simple technique for the preparation of genomic DNA from formalin-fixed, paraffin-embedded tissue. After an overnight Proteinase K digestion, genomic DNA can be manually purified from FFPE thin tissue sections in less than an hour. Amplifiable genomic DNA can be isolated from 10μm sections without centrifugation of the lysate prior to purification. Up to 12 samples can be processed in the manual format using a MagneSphere® Technology Magnetic Separation Stand (Cat.# Z5332, Z5342).
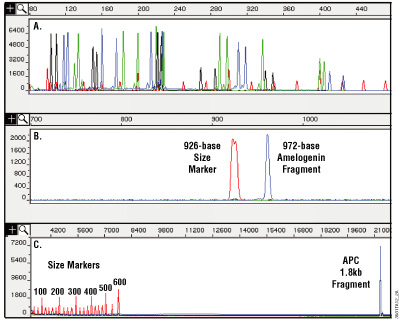
Figure 11. Analysis of DNA purified from paraffin-embedded, formalin-fixed 10µm thin sections using the MagneSil® Genomic, Fixed Tissue System. Purified DNA was amplified, and the amplification products were analyzed on an ABI PRISM® 310 or 3100 genetic analyzer. Panel A. Amplification with a set of 16 fluorescently labeled primers. Amplification products range in size from 104 to 420 bases. Panel B. A 972-base fragment amplified using an amelogenin primer set. Panel C. A 1.8kb fragment amplified from the Adenomatosis polyposis coli (APC) gene. Increasing the extension time during amplification may help to balance yields between small and large amplification products and increase yields for large amplification products. Results will vary depending on the degree of cross-linking due to formalin fixation.
One advantage this system has over other purification methods, such as phenol:chloroform extraction, is its ability to remove most inhibitors of amplification, including very small fragments of DNA. Tissue that has been stored in formalin for extended periods of time may be too cross-linked or too degraded to perform well as a template for amplification. Figure 11 shows an amplification of 16 short tandem repeat (STR) loci and demonstrates how well the isolated DNA can work in multiplex PCR using the PowerPlex® 16 HS System (Cat.# DC2101, DC2100).
The Maxwell® RSC FFPE Plus DNA Kit (Cat.# AS1720) is an automated method for purifying up to 48 samples of one to ten 5μm sections of FFPE tissue samples on the Maxwell® RSC Instrument (Cat.# AS4500; 1–16 cartridges per run) or Maxwell® RSC 48 Instrument (Cat.# AS8500; 1–48 samples per run). The FFPE Plus chemistry is designed to provide high yield of DNA from FFPE when measured by spectroscopy that is suitable for amplification applications including qPCR, multiplex PCR and NGS. The protocol provides flexibility with either a 1-hour quick deparaffinization or 24-hour overnight protocol to fit your work flow needs.
The Maxwell® RSC DNA FFPE chemistry is Promega’s latest FFPE technology and has been designed to provide highly amplifiable DNA. Save time and labor by utilizing either FFPE chemistry with the Maxwell® Instruments, and avoid exposure to hazardous xylene utilized in other FFPE purification products. Our quality testing has also demonstrated virtually no PCR inhibitors in purified DNA samples, making your PCR and other downstream applications a breeze.
Utilizing the same chemistry as the Maxwell® RSC FFPE DNA, the Maxwell® HT DNA FFPE Isolation System (Cat.# A6372) provides a simple and reliable method for high-throughput, rapid isolation of genomic DNA from FFPE tissue samples. The system does not require an organic solvent, making it safe and convenient to use, and the purified DNA can be used directly in a variety of downstream applications, including PCR and NGS.
The Maxwell® HT DNA FFPE Isolation System purifies nucleic acid using paramagnetic particles, which provide a mobile solid phase to optimize binding, washing and purification of gDNA. The use of paramagnetic particles for DNA isolation eliminates the need for centrifugation or vacuum manifolds, making the system suitable for full automation.
As FFPE samples can have widely varying quality due to the nature of the sample fixation and embedding process, QC of samples can be an important part of the FFPE workflow.



Figure 12. Comparative data of the Maxwell® RSC DNA FFPE chemistry versus the Maxwell® RSC FFPE Plus DNA chemistry. The Maxwell® RSC FFPE Plus DNA method has been observed to produce more yield by absorbance and fluorescence, while the Maxwell® RSC DNA FFPE method produces more yield by PCR.
Spectrophotometry is a common way to evaluate the quality of extracted DNA and RNA. Most laboratories have a NanoDrop Microvolume Spectrophotometer (or similar device) and they are incredibly easy to use. Pipette 1-2µl of sample, select “Analyze” and the instrument provides a read out of concentration and purity via A260/A280 and A260/A230 ratios in just a few seconds. These devices have revolutionized routine sample quantitation in the lab, but is it the best method for assessing FFPE samples? There are two main considerations when using a NanoDrop: sensitivity and integrity. FFPE samples can have a wide-ranging yield of DNA or RNA often as little as 10ng or less in a volume ranging from 10µl to 100µl from an extraction. This can result in sample concentrations below the NanoDrop’s linear range. In addition, as a spectrophotometer, it does not differentiate between RNA, DNA or free nucleotides, which can result in dramatic inaccuracies in DNA/RNA concentration measurements. Finally, there is no way to determine if a sample is accessible to downstream enzymatic assays since it cannot detect the presence or absence of crosslinks (or other damage) within a sample.
Dye-Based Quantitation like the Promega QuantiFluor® dsDNA System (Cat.# E2670, E2671), provides a rapid and significantly more sensitive method to quantitate dsDNA or RNA compared to absorbance spectroscopy. This method provides a broadly useful estimate of concentration. When considering FFPE samples, it is important to note that dye-based quantitation does not estimate the integrity of the DNA/RNA or the extent of cross-linking in the sample, which could affect success in downstream assays.
Sizing Assays (e.g., agarose gel, Tape station, fragment analyzer, DV200) can provide an estimate of concentration and—more importantly—information on the size distribution of the fragments in the sample. FFPE-derived DNA, due to the fixation process, can be significantly fragmented compared to DNA from freshly frozen tissue. Below is a fragment analyzer trace (Figure 13) and associated DV200 scores (Table 3) of DNA isolated from FFPE sections using five different purification methods. While the sizing traces do assess the distribution of DNA size purified, it does not measure the degree of cross-linking within the sample or the presence of inhibitors.
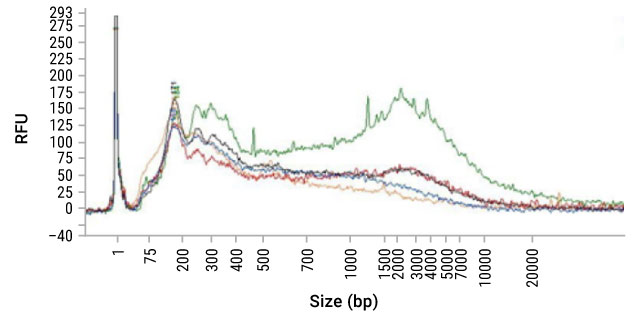
Figure 13. Fragment analyzer trace of DNA isolated from FFPE sections using five different purification methods.
Table 3. DV200 scores of DNA isolated from FFPE sections using five different purification methods in fragment analyzer trace (Figure 13).
| Method | 1 (Light Green) | 2 (Blue) | 3 (Red) | 4 (Orange) | 5 (Green) |
| DV200 | 71.8 | 69.9 | 70.1 | 58.7 | 80.5 |
For example, when the same samples were quantitated by qPCR assays of various targets and fragment sizes, the yield by qPCR does not correlate well with the DV200 scores. In fact, in this example, the samples with the lowest DV200 scores had the greatest yield by qPCR (Figure 14).



Figure 14. qPCR yields of DNA isolated from FFPE sections. The same samples of DNA isolated by five different purification methods in the fragment analyzer trace and DV200 table above were quantitated by qPCR assays of various targets and fragment sizes.
While there are general trends, the DV200 score does not necessarily correlate with success in downstream assays such as qPCR.
qPCR has several advantages for the quantitation of FFPE samples. First, qPCR can be very sensitive, requiring only a small amount of sample and detecting pg/µl amounts of DNA. In terms of sensitivity in nucleic acid detection, it is surpassed only by ddPCR. qPCR can also provide a measure of how degraded or crosslinked a DNA sample may be since nucleic acid must be a suitable substrate for a DNA polymerase for a signal to be generated. Absorbance may not represent the sample suitable for the downstream assay because it will detect DNA, fragmented DNA and nucleotides. Finally, most qPCR QC assays, such as the ProNex® DNA QC Assay (Cat.# NG1004, NG1005) provide internal controls which are used to detect the presence of inhibitors in the sample prior to attempting a more expensive assay. This can help you assess not only the integrity of the nucleic acids, but also the likelihood of an amplification-based assay to be successful.
NGS is another assay used by some labs to QC their samples. There are several reasons for this. Some labs are trying to get as much data as possible from very precious samples, in which case any sequence information may be worth the expense and risk of failed sequencing runs. As a QC test, NGS may provide a lot of information, but it is expensive and can require large amounts of sample and time. Some labs run low pass NGS, which uses highly-multiplexed samples to lower the cost per sample to determine if it is worth the time and resources to sequence deeper. Most sequencing and purification providers recommend qPCR assays to quantitate FFPE DNA, as all NGS workflows depend primarily on the success of enzymatic amplification steps to obtain sequencing-ready DNA as part of library preparation steps.Table 4. Comparative Pros and Cons of Various QC Assays.
| Method | Speed | Sensitivity | Quantitative? | Measure Purity? | Assess size of NA? | Detect Cross- linking? | Cost |
|---|---|---|---|---|---|---|---|
| Spectrophotometry (NanoDrop) | +++ | + | Semi - quantitative | + | - | - | $ |
| Dye-Based Quantitation | +++ | ++ | Y | - | - | - | $$ |
| DV200 | ++ | + | Semi - quantitative | - | +* | - | $$$ |
| Gel Electrophoresis | ++ | +/- | Semi - quantitative | - | +* | - | $$ |
| qPCR | ++ | +++ | Y | +* | + | + | $$ |
| NGS | + | Y | + | ++ | + | $$$$$ |
Plant Genomic DNA Isolation
The Wizard® Magnetic 96 DNA Plant System (Cat.# FF3760, FF3761) is designed for manual or automated 96-well purification of DNA from plant leaf and seed tissue. The Wizard® Magnetic 96 DNA Plant System has been validated with corn and tomato leaf as well as with canola and sunflower seeds. The DNA purified from these samples can be used in PCR and other more demanding applications, such as RAPD analysis. Since plant materials can be particularly challenging to lyse, especially when working with tough or woody tissues, additional required equipment includes not only a magnet (MagnaBot® FLEX 96 Magnetic Separation Device, Cat.# VA1920) but also a device capable of breaking up seed or leaf material (e.g., Geno/Grinder® 2000 from SPEX CertiPrep, Inc.).
The yield depends on the source material and how well the seeds or leaf disks are pulverized prior to the genomic DNA isolation. Yield may range from 10–100ng from a single 8mm leaf punch. To increase the yield from the Wizard® Magnetic 96 DNA Plant System, a scale up in volume with up to 5 leaf punches can be used [as demonstrated in Promega Notes 79]. The potential scale-up is limited by the volume in a deep-well, 96-well plate.
Another automated option we have to meet your plant DNA extraction needs, is the Maxwell® RSC Plant DNA Kit (Cat.# AS1490). The Maxwell® RSC Plant DNA Kit is used with the Maxwell® RSC and RSC 48 Instruments to provide an easy method for efficient, automated purification of genomic DNA (gDNA) from a range of plant tissue samples, including corn, soybean and Arabidopsis. The Instruments are supplied with preprogrammed purification methods and uses predispensed reagent cartridges, maximizing simplicity and convenience.
Using this system, DNA can be purified from plant samples in under 60 minutes with minimal preprocessing and no organic extractions. Automated purification results in consistent purification, with less variability than traditional DNA extraction methods such as CTAB and spin-columns. The resulting purified DNA is ready to use in downstream applications, including amplification assays.
Serum-Plasma Genomic DNA Isolation
For high quality, purified cell-free DNA from plasma samples, we offer the Maxwell® RSC ccfDNA Plasma Kit (Cat.# AS1480). Utilizing the simple three-step protocol, the Maxwell® RSC Instrument can process 1 to 16 samples, and the Maxwell® RSC 48 Instrument can process 1 to 48 samples. Simply add 0.2–1.0ml of plasma to the prepared cartridges and select Start, no preprocessing of samples required. In approximately 70 minutes, you will have high yields of amplifiable DNA that is ready to be used in downstream assays including qPCR, NGS and digital PCR.
As a magnetic particle mover, not a liquid handler, the Maxwell® RSC additionally offers several advantages over other automated systems. Since no liquid handling or splashing occurs during sample processing, there is minimal risk of sample cross-contamination. It also eliminates the worry of potential clogs and inevitable system breakdowns that follow, ensuring a smooth workflow with fewer disruptions.
Bacterial Genomic DNA Isolation
If you need to make quick decisions about potential food contamination and spoilage, the Maxwell® RSC PureFood Pathogen Kit (Cat.# AS1660) offers a simple automated protocol with minimal hands-on steps. The kit effectively eliminates laborious sample preprocessing steps such as enzymatic pretreatment, as it works with inhibiting sample types and also has the ability to lyse both Gram+ or Gram– bacteria.
By coupling the high-performance Maxwell® chemistries with the trusted benchtop Maxwell® RSC instruments, you will be able to effectively purify bacterial DNA from up to 48 food samples in as little as 40 minutes. Once extracted, the resulting DNA is ready for advanced downstream molecular analyses, including serotyping, NGS and identification of spoilage organisms.
This method can be utilized for both raw and processed food and has successfully been used to isolate pathogen DNA from a wide variety of food samples, including E. coli 0157:H7 from uncooked beef, Salmonella enterica from uncooked chicken and Listeria monocytogenes from whole milk. Figure 15 below highlights a comparison of total DNA versus E. coli 0157:H7 DNA extracted from cilantro samples that were spiked with the E. coli 0157:H7 bacteria.

Figure 15. Comparison of total DNA and E. coli 0157:H7 DNA extracted from cilantro samples spiked with the indicated amounts of E. coli 0157:H7 bacteria. The total DNA concentration was assessed using the QuantiFluor® ONE dsDNA System.
Buffy Coat Genomic DNA Isolation
The Maxwell® RSC Buffy Coat DNA Kit (Cat.# AS1540) provides a simple, automated method of genomic DNA extraction using the convenient, prefilled cartridge format of the Maxwell® RSC Instrument. The kit contains all the reagents you need for optimal DNA extraction, and is compatible with blood stored in EDTA, heparin and citrate anticoagulants. Avoid the tedious and time-consuming hassle of preprocessing samples, simply add 50–250μl of your sample directly into well #1 of the cartridges. Your purified DNA is ready for analysis in about 50 minutes, and can be used directly in various downstream applications, such as agarose gel electrophoresis.
Food Genomic DNA Isolation
Food and plant materials often provide the greatest challenge for cell lysis and intact DNA extraction, due to the lysis conditions required to liberate the nucleic acid and the processing of plant materials into comestibles.
Another specialized genomic DNA isolation system is the Wizard® Magnetic DNA Purification System for Food (Cat.# FF3750, FF3751). This convenient protocol is designed for the manual purification of DNA from a variety of food samples including corn seeds, cornmeal, soybeans, soy flour and soy milk, generating results in one-third of the time of traditional methods. In addition, DNA can be purified from processed food such as corn chips, chocolate and chocolate-containing foods, lecithin and vegetable oils if used with the appropriate optimized protocols.
The DNA purified from many of these samples can be used in PCR-based testing for Genetically Modified Organism (GMO) DNA sequences, such as by quantitative analysis using TaqMan® assays. As with all isolation systems using the MagneSil® PMPs, a magnetic separation stand is needed and enables processing of up to 12 samples per batch. With samples containing highly processed food, the genomic DNA isolated will be fragmented and better suited for analysis using amplification rather than a Southern blot. The yield of DNA from this system will vary depending on source type and extent of food processing.
The Maxwell® RSC PureFood GMO and Authentication Kit (Cat.# AS1600) provides an easy and automated method for efficient purification of DNA for PCR-based food and ingredient authentication. The Kit is used with the Maxwell® RSC and RSC 48 Instruments and can purify DNA from raw and processed food samples, including corn, soybeans, canola, ground beef and ground pork.
The five-step, ~100 minute protocol requires only 30 minutes of hands-on time, effectively achieving not only faster results with walk-away automation, but also freeing up laboratory resources for higher value activities. The purified DNA extracted using the PureFood Kit is ready to be used for several applications, including real-time PCR, gel electrophoresis, next-generation and Sanger sequencing and microarrays.
Nucleic Acid Purification Protocols for Plant & Food Sample Types
Explore our collection of protocols for manual and automated DNA or RNA extraction from a variety of food and plant samples.
Plasmid DNA Purification
The primary consideration for plasmid purification is separation of plasmid DNA from the chromosomal DNA and cellular RNA of the host bacteria. A number of methods have been developed to generate a cleared lysate that not only remove protein and lipids, but also efficiently remove contaminating chromosomal DNA while leaving plasmid DNA free in solution. Methods for the preparation of cleared lysates that enrich for plasmid DNA include SDS-alkaline denaturation (22–23), salt-SDS precipitation (24) and rapid boiling (25).
The SDS-alkaline denaturation method, which is used in all Promega plasmid isolation systems, is a popular procedure for purifying plasmid DNA because of its overall versatility and consistency. This technique exploits the difference in denaturation and renaturation characteristics of covalently closed circular plasmid DNA and chromosomal DNA fragments. Under alkaline conditions (at pH 11), both plasmid and chromosomal DNA are efficiently denatured. Rapid neutralization with a high-salt buffer such as potassium acetate in the presence of SDS has two effects that contribute to the overall effectiveness of the method.
First, rapid neutralization causes the chromosomal DNA to base-pair in an intrastrand manner, forming an insoluble aggregate that precipitates out of solution. The covalently closed nature of the circular plasmid DNA promotes interstrand rehybridization, allowing the plasmid to remain in solution. Second, the potassium salt of SDS is insoluble, so the protein and detergent precipitate and aggregate, which assists in the entrapment of the high-molecular-weight chromosomal DNA. Separation of soluble and insoluble material is accomplished by a clearing method (e.g., filtration, magnetic clearing or centrifugation). The soluble plasmid DNA is ready to be further purified. There are several methods available to purify plasmid DNA from cleared lysate. These include:
- binding plasmid to silica in the presence of high concentrations of chaotropic salts (2–4)
- differential precipitation of plasmid DNA from aqueous chaotropic salt/ethanol solutions (26–28)
- ion exchange chromatography over DEAE-modified cellulose membranes (29)
- precipitation with polyethylene glycol (30–31) organic extraction using phenol (32)
General Considerations for Plasmid DNA Purification
Bacterial Growth and Culture Conditions
Successful isolation of quality plasmid DNA begins with culture preparation. A number of factors can influence the growth of bacterial cells. Bacterial growth in liquid culture occurs in three phases: 1) a short lag phase in which the bacteria become acclimated to the media and begin to divide; 2) a log phase, characterized by exponential growth in which most strains of E. coli will divide every 20–30 minutes; and 3) a stationary phase in which growth slows and eventually stops in response to the lack of nutrients in the medium.
No net increase in biomass will occur in the stationary phase, but plasmid replication will continue for several hours after reaching stationary phase. Most strains of E. coli will reach a concentration of 1.0–4.0 × 109 cells/ml of culture at this stage, depending on culture media and aeration conditions. Depending on inoculation size and the size of the culture, stationary phase will be reached in 6–8 hours
Aeration and temperature are of critical importance. The culture volume should be less than or equal to 1/4 the volume of the container (e.g., 250ml medium in a 1 liter flask); using 1/10 the container volume (e.g., 100ml medium in a 1,000ml flask) produces optimal results. The culture tube or flask should be placed in an orbital shaker (approximately 250rpm) to ensure adequate aeration (33). Baffled flasks may increase aeration and thus yields of plasmid DNA. Since most strains of E. coli grow best at 37°C, this incubation temperature is recommended unless the strain of interest requires different conditions for optimal growth.Different culture media will also have a profound effect on the growth of different bacterial strains. Promega plasmid DNA purification systems are appropriate for bacterial cultures grown in 1X Luria-Bertani (LB) medium. However, use of LB-Miller medium containing more NaCl will produce significantly greater yields and is highly recommended. Richer media such as 2X YT, CIRCLEGROW® or Terrific Broth may be used to increase plasmid yields by increasing the biomass for a given volume of culture.
Keep the biomass in a range acceptable for the plasmid isolation system used, as overloading may result in poor purity and yield of the plasmid DNA (see Biomass Processed for more information). Culture incubation time affects both the yield and quality of plasmid DNA isolated. Bacterial cultures grown to insufficient density will yield relatively low amounts of DNA. Overgrown cultures may result in suboptimal yields and excessive chromosomal DNA contamination due to autolysis of bacterial cells after they have reached stationary phase. We do not recommend the use of cultures grown longer than 18–20 hours.
Antibiotic Selection
Most plasmids carry a marker gene for a specific antibiotic resistance. By supplementing the growth medium with the antibiotic of choice, only cells containing the plasmid of interest will propagate. Adding antibiotic to the required concentration will help to maximize plasmid yields. Note that adding too much antibiotic can inhibit growth, and too little may cause a mixed population of bacteria to grow—both with and without the plasmid of interest. For more information on optimal antibiotic ranges to use in culture as well as the mechanisms of antibiotic action and resistance, see Table 5 (34).
Table 5. Antibiotic Mode of Action and Mechanism of Resistance.
| Antibiotic | Mode of Action | Mechanism of Resistance | Working Conc. | Stock Solution |
|---|---|---|---|---|
| Ampicillin (Amp) | A derivative of penicillin that kills growing cells by interfering with bacterial cell wall synthesis. | The resistance gene (bla) specifies a periplasmic enzyme, β-lactamase, which cleaves the β-lactam ring of the antibiotic. | 50–125µg/ml | 50mg/ml in water |
| Chloramphenicol (Cm) | A bacteriostatic agent that interferes with bacterial protein synthesis by binding to the 50S subunit of ribosomes and preventing peptide bond formation. | The resistance gene (cat) specifies an acetyltransferase that acetylates, and thereby inactivates, the antibiotic. | 20–170µg/ml | 34mg/ml in ethanol |
| Hygromycin (Hygro) | A protein synthesis inhibitor that interferes with 80S ribosome translocation and causes mistranslation. | The resistance gene (hph) specifies a phosphotransferase that catalyzes the phosphorylation of the 4-hydroxyl group on the cyclitol ring (hyosamine), thereby producing 7′-O-phosphoryl-hygromycin B, which lacks biological activity both in vivo and in vitro. | 20–200µg/ml | 100mg/ml in water |
| Kanamycin (Kan) | A bactericidal agent that binds to 70S ribosomes and causes misreading of messenger RNA. | The resistance gene (kan) specifies an enzyme (aminoglycoside phosphotransferase) that modifies the antibiotic and prevents its interaction with ribosomes. | 30µg/ml | 50mg/m in water |
| Neomycin (Neo) | A bactericidal agent that blocks protein synthesis by binding to the prokaryotic 70S ribosomal subunit. | Expression of the bacterial APH (aminoglycoside phosphotransferase) gene (derived from Tn5). | 50µg/ml | 25mg/ml in water |
| Tetracycline (Tet) | A light-sensitive bacteriostatic agent that prevents bacterial protein synthesis by binding to the 30S subunit of ribosomes. | The resistance gene (tet) specifies a protein that modifies the bacterial membrane and prevents transport of the antibiotic into the cell. | 10µg/ml in liquid culture; 12.5µg/ml in plates | 12.5mg/ml in ethanol |
Recommended Inoculation Procedures
1–100ml of Culture
Pick an isolated colony from a freshly streaked plate (less than 5 days old) and inoculate LB medium containing the required antibiotic(s). Incubation with shaking for 8–16 hours at 37°C before harvesting generally results in maximum yields of a high-copy-number plasmid. To achieve a highly reproducible yield, determine the cell density reached in a typical experiment, and grow cultures to this density in each subsequent experiment. Typically, after overnight incubation, the absorbance of a tenfold dilution of the culture at a wavelength of 600nm (A600) with a 1cm path length should range from 0.10–0.35.
100–1000ml of Culture
Using a colony from a freshly streaked plate (less than 5 days old), inoculate 5–50ml of LB medium containing the required antibiotic(s). Grow this starter culture from 8 hours to overnight at 37°C. The following day, use this culture to inoculate the larger culture flask containing antibiotic-supplemented medium by diluting the starter culture between 100- to 500-fold (e.g., adding 10ml overnight culture to 1 liter medium). Incubate this secondary culture for 12–16 hours before harvesting cells. The A600 of a tenfold dilution of the culture should be 0.10–0.35. As with smaller cultures, to achieve a highly reproducible yield, determine the cell density used in a typical experiment and grow cultures to this density in each subsequent experiment.
Harvesting
When harvesting bacteria, follow the conditions outlined in either the Wizard® Plus SV Miniprep DNA Purification System or the PureYield™ Plasmid Midiprep System protocol. If the recommended centrifugation time or speed is exceeded, the pelleted cells may be more difficult to resuspend. Insufficient centrifugation time or speed may result in incomplete harvesting of cells and loss of starting material. Consult a centrifuge instruction manual for conversion of rpm to g-force. Once the bacteria are pelleted, this is a good stopping point in the purification process. Storing the pellet at –20°C results in little loss of plasmid DNA and may enhance lysis.
Factors That Affect Plasmid DNA Quality and Yield
Bacterial Strain Selection
The choice of host bacterial strain can have a significant impact on the quality and yield of DNA using any purification method. We recommend the use of host strains such as DH5α™, JM109 (Cat.# L2005) and XL1-Blue, which contain mutations in the endA gene. E. coli strains that are listed as endA1 contain such mutations.
The endA gene encodes a 12kDa periplasmic protein called endonuclease I. This enzyme is a double-stranded DNase that can copurify with plasmid DNA, thus causing potential degradation. RNA acts as a competitive inhibitor and alters the endonuclease specificity from that of a double-stranded nucleolytic enzyme yielding seven-base oligonucleotides to a nickase that cleaves an average of one time per substrate (35–36). The function of endonuclease I is not fully understood, and strains bearing endA1 mutations have no obvious phenotype other than improved stability and yield of plasmid obtained from them.
The expression of endonuclease I has been characterized and was found to be dependent on bacterial growth phase (37). In this study, endonuclease I levels were found to be more than 300 times higher during exponential phase compared to stationary phase. In addition, media compositions that encouraged rapid growth (e.g., high glucose levels and addition of amino acids) resulted in high endonuclease I levels.
Strains that contain the wildtype endonuclease A (endA) gene can yield high-quality, undegraded plasmid DNA if special precautions are used to reduce the probability of nuclease contamination and plasmid degradation (37). Promega has performed a thorough investigation of methods at different points in the purification process to ensure the isolation of high-quality DNA from EndA+ (wildtype) bacterial strains. These include: 1) inclusion of an alkaline protease treatment step that degrades nucleases in the Wizard® Plus SV Minipreps DNA Purification System; 2) optimization of culture conditions to limit in vivo expression during bacterial growth; 3) heat inactivation during and after purification; 4) optimization of protocol conditions to limit binding of the nuclease to the resin and 5) post-purification methods to remove endonuclease. These methods and results are summarized in Schoenfeld et al. 1995 (38) and the Wizard® Plus SV Plasmid DNA Purification System Technical Bulletin. Information on genetic markers in bacterial strains can also be found in Ausubel et al. 1989 (33) and Sambrook et al. 1989 (39).
Plasmid Copy Number
One of the most critical factors affecting the yield of plasmid from a given system is the copy number of the plasmid. Copy number is determined primarily by the region of DNA surrounding and including the origin of replication in the plasmid. This area, known as the replicon, controls replication of plasmid DNA by bacterial enzyme complexes. Plasmids derived from pBR322 (Cat.# D1511) contain the ColE1 origin of replication from pMB1. This origin of replication is tightly controlled, resulting in approximately 25 copies of the plasmid per bacterial cell (low copy number). Plasmids derived from pUC contain a mutated version of the ColE1 origin of replication, which results in reduced replication control and approximately 200–700 plasmid copies per cell (high copy number).
Some plasmids contain the p15A origin of replication, which is considered a low-copy-number origin. The presence of the p15A origin of replication allows for replication of that particular plasmid in conjunction with a plasmid containing the ColE1 origin of replication. A compatibility group is defined as a set of plasmids whose members are unable to coexist in the same bacterial cell. They are incompatible because they cannot be distinguished from one another by the bacterial cell at a stage that is essential for plasmid maintenance. The introduction of a new origin, in the form of a second plasmid of the same compatibility group, mimics the result of replication of the resident plasmid. Thus, any further replication is prevented until after the two plasmids have been segregated to different cells to create the correct prereplication copy number (40). Most plasmids provided by Promega, including the pGEM® Vectors, are considered to be high-copy-number. The only exception is the pALTER®-MAX Vectors.
Some DNA sequences, when inserted into a particular vector, can lower the copy number of the plasmid. Furthermore, large DNA inserts can also reduce plasmid copy number. In many cases, the exact copy number of a particular construct will not be known. However, many of these plasmids are derived from a small number of commonly used parent constructs.
Appropriate Sample Size and Throughput
Depending on the volume of the bacterial culture, there are different isolation systems for your needs. For small-volume bacterial cultures of 0.6–3ml, use a system like the PureYield™ Plasmid Miniprep System (Cat.# A1223, A1222), which gives a plasmid DNA yield of 1.5–7.5μg with an A260/A280 ≥1.8 from a 0.6ml overnight bacterial culture with a total biomass (O.D.600 of culture × volume of culture in μl) of 1.3–8.

Figure 16. The Vac-Man® 96 Vacuum Manifold. This 96-well vacuum manifold is used for processing SV 96 plates for plasmid, genomic and PCR product purification.
For larger cultures with volumes ranging from 50–100ml, the PureYield™ Plasmid Midiprep System (Cat.# A2492, A2495, A2496) is a good choice. With this system, a 50ml culture of a high-copy-number plasmid with a total biomass of 100–200 O.D.600 units will yield 100–200µg of plasmid. The PureYield™ Plasmid Maxiprep System (Cat.# A2392, A2393) can isolate plasmid from 100–250ml of culture with yields up to 1mg of plasmid DNA with an A260/A280 >1.7 from 250ml of overnight bacterial culture, transformed with a high-copy-number plasmid.
For high-throughput processing, systems based on a 96-well format can be performed manually with a vacuum manifold (e.g., Vac-Man® 96 Vacuum Manifold; Figure 16) using silica membrane technology such as the Wizard® SV 96 Plasmid DNA Purification System (Cat.# A2250, A2255, A2258). Alternatively, an automated liquid-handling workstation can process multiwell plates with MagneSil® PMPs and a 96-well magnet (e.g., MagnaBot® 96 Magnetic Separation Device; Figure 17) using the Wizard® MagneSil® Plasmid Purification System (Cat.# A1630, A1631, A1635).

Figure 17. The MagnaBot® 96 Magnetic Separation Device. This 96-well magnet is used for capturing MagneSil® PMPs for DNA purification.
Yields for these systems using high-copy-number plasmid range from 3–5µg for the Wizard® SV 96 Plasmid DNA Purification System and up to 6µg for the Wizard® MagneSil® Plasmid Purification System. Smaller plasmid amounts are helpful for assessing the success of a cloning experiment by PCR or restriction digestion or for use in a coupled transcription/translation system like the TNT® Quick Coupled Transcription/Translation System (Cat.# L1170, L2080).
Biomass Processed
Optical density (O.D.) is the measure of how much light is blocked by the biomass of the bacterial culture in a path length of 1cm. The density of the culture is measured at a wavelength of 600nm and can have a great effect on plasmid isolation success. For example, the Wizard® SV 96 Plasmid Purification System has a maximum biomass recommendation of 4.0 O.D.600 to avoid clogging of the Wizard® SV 96 Lysate Clearing Plate (Cat.# A2241, A2248), so calculating the O.D. of the culture is necessary.
O.D./ml culture = 600nm absorbance reading × dilution factor
For O.D. measurement, a 1:10 dilution is typically used (e.g., 0.1ml culture in 0.9ml culture medium) to keep the reading in the range of 0.1–1.0, where the spectrophotometer is most accurate. For the example above, if the 1:10 dilution reading is 0.15, meaning that each milliliter of culture is 1.5 O.D., no more than 2.67ml culture can be processed (4 O.D. divided by 1.5 O.D./ml = 2.67ml). Exceeding the recommendations of the plasmid purification system may cause clogging or contamination of the system.
Plasmid Purification Method and Transfection
Many plasmid isolation systems indicate they are transfection-quality (e.g., the PureYield™ Plasmid Systems or the Wizard MagneSil Tfx™ System, Cat.# A2380). This may be important, as some cultured cells are sensitive to the amount of endotoxin and other contaminants present in the plasmid preparation. Endotoxin is a lipopolysaccharide cell wall component of the outer membrane of Gram-negative bacteria (i.e., all E. coli strains) that can copurify with the plasmid DNA regardless of the purification system used. The amount of this molecule varies by bacterial strain, growth conditions and isolation method. In the PureYield™ Plasmid Systems, there is an Endotoxin Removal Wash solution that reduces the amount of endotoxin, proteins and other contaminants eluted with the plasmid DNA. For many common cell lines, like 293 and HeLa, the amount of endotoxin present for routine transfections has a minimal effect on the efficiency of transfection (41).
Many factors influence transfection efficiency and/or cellular death including the type and amount of transfection reagent, cell confluency, DNA amount and incubation time with the reagent:DNA complex. Each of these factors will need to be optimized for each cell line-plasmid combination transfected in order to minimize cell death and maximize transfection efficiency. In our experience, transfection experiments with HeLa and NIH/3T3 cells demonstrated that there was little DNA preparation difference with four different plasmid isolation systems used (based on silica membrane, anion exchange and silica resin) when comparing efficiencies using the same transfection reagent. However, the transfection reagent used for DNA uptake had a significant effect on transfection efficiency and cell death. For general considerations for optimization, consult our Transfection guide.
Silica Column-Based Systems
Promega products like the Wizard® Plus SV Minipreps DNA Purification System (Cat.# A1330, A1460, A1465) and the PureYield™ Plasmid Systems combine the benefits of alkaline lysis with the rapid and easy purification by silica. This is done by using a silica-based membrane in a column format to bind the plasmid DNA contained in the cleared alkaline lysates. Purification is based on selective adsorption of DNA to the silica membrane in the presence of high concentrations of chaotropic salts, washes to efficiently remove contaminants, and elution of the DNA with low-salt solutions such as TE buffer or water.
Purified plasmid DNA is used in many applications from preparing vectors for cloning to generating templates for transcription or coupled transcription/translation reactions. The silica-based purification systems from Promega minimize the amount of salts and other impurities carried over during isolation, which can negatively affect downstream applications, lower yield or prevent enzyme systems from synthesizing the product of interest.
PureYield™ Plasmid Systems
The PureYield™ Plasmid Systems isolates high-quality plasmid DNA for use in eukaryotic transfection and in vitro expression experiments. The unique reagents, proprietary matrix and silica membrane-based design of the PureYield™ Systems greatly reduces the amount of time spent on purification compared to silica resin or other membrane-column methods. While the unique Endotoxin Removal Wash removes protein, RNA and endotoxin contaminants from the bound DNA, the Column Wash Solution followed by membrane drying eliminates salts and alcohols from the plasmid prep, allowing the purified plasmid to be used for highly sensitive applications such as transfection, in vitro transcription and coupled in vitro transcription/translation. An additional benefit is that the same degree of purification can be obtained even with low-copy-number plasmids. Although the system works best for plasmids less than 10kb, plasmids as large as 18kb have been purified.
The unique combination of reagents in the PureYield™ Plasmid Miniprep System purifies plasmid either directly from 0.6ml of bacterial culture or cell pellets from up to 3ml of cell culture (Figure 18). A typical overnight culture is grown in LB medium for 16–18 hours. If the cell pellet method is chosen, cells are harvested by centrifugation, then resuspended in 600μl of TE buffer or water. Purifying DNA directly from bacterial culture takes less than 10 minutes with elution volumes as low as 30μl, resulting in more concentrated plasmid DNA. The low elution volume is possible because the column design retains virtually no buffer. A transfection comparison of plasmid isolated using the PureYield™ Plasmid Miniprep System in various cell lines can be found in Figure 19.
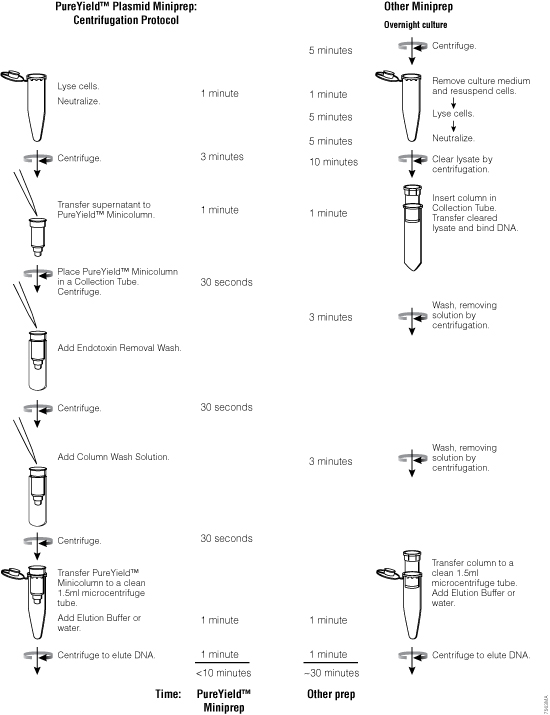
Figure 18. The PureYield™ Plasmid Miniprep System yields transfection-quality DNA in approximately 10 minutes.
To isolate larger quantities of high-quality plasmid DNA, use the PureYield™ Plasmid Midiprep System. This plasmid midiprep system is designed to purify 100–200µg of plasmid DNA with an A260/A280 >1.7 from a 50ml overnight culture of bacteria in as little as 30 minutes, if the culture is grown with a high-copy-number plasmid, reaching a total optical density (O.D.600 of culture × volume of culture) of 100–200. Larger volumes up to 250ml can be processed, but require greater volumes of solutions than that supplied with the PureYield™ Plasmid Midiprep System.
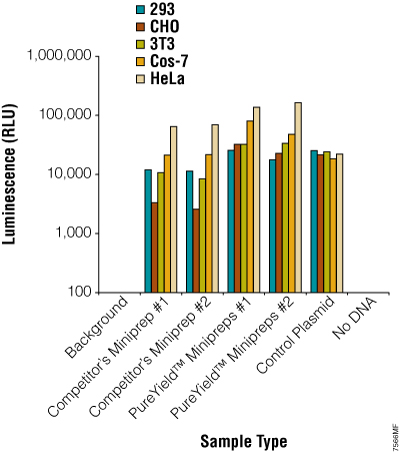
Figure 19. Plasmid DNA prepared using the PureYield™ Plasmid Miniprep System consistently works well in transfection experiments. The pGL4.13[luc2/SV40] Vector (Cat.# E6681) was prepared using a competing system or the PureYield™ Plasmid Miniprep System. Five different commonly used mammalian cell lines were transfected with the plasmid, and transfection efficiency was assessed by measuring the luciferase activity using the ONE-Glo™ Luciferase Assay System (Cat.# E6110; n = 6).
The PureYield™ Plasmid Midiprep System is designed for purification by vacuum using a manifold such as the Vac-Man® Laboratory Vacuum Manifold (Cat.# A7231), but there are alternative protocols that use all centrifugation or both vacuum and centrifugation. All protocols generate high-quality purified plasmid DNA. A swinging-bucket tabletop centrifuge or the Eluator™ Vacuum Elution Device (Cat.# A1071) is required for the final elution step regardless of the protocol chosen.
For a larger plasmid isolation capacity, the PureYield™ Plasmid Maxiprep System is able to purify up to 1mg of plasmid DNA with an A260/A280 >1.7 from 250ml of overnight bacterial culture, transformed with a high-copy-number plasmid in approximately 60 minutes. As with the midiprep system, the protocol requires a vacuum pump and manifold (e.g., the Vac-Man® Laboratory Vacuum Manifold, 20-sample), a centrifuge with a fixed-angle rotor for lysate clearing and either a tabletop centrifuge with a swinging bucket rotor or the Eluator™ Vacuum Elution Device for the final elution step.
Wizard® SV Column-Based Systems
High-quality, purified plasmids are used for automated fluorescent DNA sequencing as well as for other standard molecular biology techniques including restriction enzyme digestion and PCR. Whether you are isolating a few samples or a 96-well plate, there is a silica membrane-based system available.
For manual purification, the Wizard® Plus SV Minipreps DNA Purification System provides a simple and reliable method for rapid isolation of plasmid DNA using a column-based silica membrane (see Figure 20 for overview of method). The entire miniprep procedure can be completed in 30 minutes or less, depending on the number of samples processed. The plasmid DNA from 1–10ml of overnight E. coli culture can be purified by using either a vacuum manifold like the Vac-Man® Laboratory Vacuum Manifold (process up to 20 samples) or a microcentrifuge (number of samples processed depends on rotor size).
This system can be used to isolate any plasmid hosted in E. coli but works most efficiently when the plasmid is less than 20,000bp in size. The yield of plasmid will vary depending on a number of factors, including the volume of bacterial culture, plasmid copy number, type of culture medium and the bacterial strain used as discussed in Factors that Affect Plasmid DNA Quality and Yield. The DNA binding capacity of the SV membrane is up to 20µg of high-quality plasmid DNA. An alkaline protease treatment step in the isolation procedure improves plasmid quality by digesting proteins like endonuclease I.
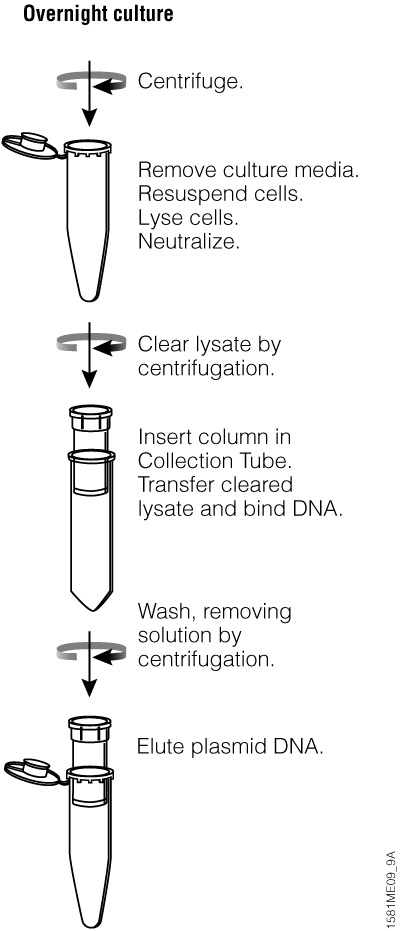
Figure 20. Overview of the Wizard® Plus SV Minipreps DNA Purification System centrifugation protocol.
To process more samples at once, consider using the 96-well format of the Wizard® SV 96 and SV 9600 Plasmid DNA Purification Systems. These high-throughput systems provide a simple and reliable method for the rapid isolation of plasmid DNA using a silica-membrane 96-well plate. A single plate can be processed in 60 minutes or less. The Wizard® SV 96 and SV 9600 Systems are designed for use either in a manual format or with automated instruments.
To use the Wizard® SV 96 and SV 9600 Systems, a vacuum manifold (e.g., Vac-Man® 96 Vacuum Manifold) and a vacuum pump capable of generating 15–20 inches of mercury or equivalent with a vacuum trap is needed for sample processing. Figure 14 shows the Vac-Man® 96 Manifold set up for purification.
Magnetic-Based
For automated, high-throughput plasmid purification, use our MagneSil® paramagnetic particle (PMP)-based systems that yield purified plasmid, which can be used directly for automated fluorescent DNA sequencing, as well as for other standard molecular biology techniques including restriction enzyme digestion and PCR.
Ideal for use with automated platforms, the silica-coated MagneSil® PMP systems are also easily scalable for larger volumes or multiwell format. For plasmid miniprep purification, the MagneSil® PMPs are used for both lysate clearing and DNA binding, eliminating the need for centrifugation or vacuum filtration, as the binding of nucleic acids occurs in solution. The particles are also completely resuspended during the wash steps of a purification protocol, enhancing the removal of impurities from the DNA.
The Wizard® MagneSil® Plasmid DNA Purification System provides a simple and reliable method for the rapid isolation of plasmid DNA in a multiwell format. The purification procedure uses MagneSil® PMPs for lysate clearing as well as DNA capture, circumventing the need for centrifugation or vacuum filtration. The MagnaBot® 96 Magnetic Separation Device is needed for plasmid purification. The protocol also requires a multiwell plate shaker. This protocol has been optimized using the Micro Mix 5 shaker on the Beckman Coulter Biomek® 2000.
The Wizard MagneSil Tfx™ System provides a simple and reliable method for the rapid isolation of transfection-quality plasmid DNA in a multi-well format. DNA purified with using this system is greatly reduced in chemical contaminants as well as RNA, protein, and endotoxin, providing high-quality plasmid DNA suitable for transfection, as well as for other standard molecular biology techniques. Like the Wizard® MagneSil® Plasmid DNA Purification System, the Wizard MagneSil Tfx™ System uses MagneSil® PMPs for lysate clearing as well as DNA capture. In addition, a proprietary paramagnetic endotoxin removal resin reduces the level of endotoxin present in the purified plasmid DNA.
By avoiding the need for centrifugation or vacuum filtration, DNA purification with the Wizard MagneSil Tfx™ System can be completely automated, requiring the MagnaBot® 96 Magnetic Separation Device and Heat Transfer Block for the protocol. An automated method for the Wizard MagneSil Tfx™ System has been developed for the Biomek® FX robotic workstation. The procedure requires no manual intervention and takes approximately 45 minutes to process a single 96-well plate. This automated protocol also can be adapted to other robotic workstations.
Plasmid Purification Kit Comparison
Compare plasmid DNA prep kits to find the purification solution that is right for you.
DNA Fragment Purification from Agarose Gels and PCR Amplification
PCR & Gel Clean-Up and Concentration
Purification of DNA fragments or PCR products does not involve disruption of cellular structures in order to liberate DNA, but rather separation of DNA from in vitro reactions or agarose gel slices. After a PCR amplification or restriction enzyme digestion, the reaction components include protein and salts that may inhibit subsequent applications and will need to be removed from the DNA fragments. An agarose gel may be run to isolate a fragment of the correct size if there is more than one product present. Fragment DNA purification can improve efficiency in subsequent reactions.
For example, it’s often the case that PCR products can be used directly in T-vector cloning. However, nonspecific amplification products and primer dimers can compete for ligation with the desired PCR product, resulting in a low frequency of positive clones. Additionally, removing the reaction components prior to sequencing will ensure the right primers are used for sequencing reactions and that the fluorescently labeled nucleotides are not competing with the unlabeled dNTPs remaining from the PCR amplification.
Applications such as cloning, labeling and sequencing DNA frequently require the purification of DNA fragments from agarose gels or amplification reactions. Promega provides multiple systems for DNA fragment purification, including three based on silica membrane technology (ReliaPrep™ Clean-Up and Concentration System, Wizard® SV Gel and PCR Clean-Up System and Wizard® SV 96 PCR Clean-Up System) and one based on MagneSil® PMPs (Wizard® MagneSil® Sequencing Reaction Clean-Up System).
ReliaPrep™ DNA Clean-Up and Concentration System
The ReliaPrep™ Clean-Up and Concentration System (Cat.# A2891, A2892, A2893) is designed to quickly concentrate and purify dilute DNA solutions, extract and purify DNA fragments of 100bp–10kb from standard or low-melt agarose gels or to purify products directly from a PCR amplification. This membrane-based system can bind up to 60µg of DNA and concentrate as much as 300µl of dilute DNA, recovering isolated DNA fragments or PCR products in as little as 10 minutes, depending on the number of samples processed and the protocol used. Up to 96% recovery is achieved, depending on starting DNA size (Table 6). A single reagent stream is used for all three procedures, making the system both fast and easy. The purified DNA can then be used for automated fluorescent DNA sequencing, cloning, labeling, restriction enzyme digestion, NGS or in vitro transcription/translation.
Table 6. Percent Recovery Versus Starting DNA Using the ReliaPrep™ DNA Clean-Up and Concentration System.
| Starting DNA | Percent Recovery |
|---|---|
| 1µg | 96% |
| 3µg | 95% |
| 5µg | 89% |
| 10µg | 84% |
Wizard® SV Gel and PCR Clean-Up System
The Wizard® SV Gel and PCR Clean-Up System (Cat.# A9281, A9282, A9285) provides a reliable method to purify double-stranded, PCR-amplified DNA either directly from the reaction or from agarose. The system is designed to extract and purify DNA fragments of 100bp to 10kb from standard or low-melting point agarose or to purify PCR products directly from an amplification reaction, using the SV silica membrane column. This purification kit is a single column system that can be used with a vacuum manifold (e.g., Vac-Man® Laboratory Vacuum Manifold or a standard microcentrifuge). Up to 95% recovery is achieved, depending upon the DNA fragment size (see Table 7). PCR products are commonly purified to remove excess nucleotides, primers and PCR additives like DMSO and betaine (Table 8). This membrane-based system, which can bind up to 40µg DNA, allows recovery of isolated DNA fragments or PCR products in as little as 20 minutes, depending on the number of samples processed and the protocol used. The purified DNA can be used for automated fluorescent DNA sequencing, cloning, labeling, restriction enzyme digestion or in vitro transcription/translation without further manipulation.
Table 7. Percent Recovery Versus Double-Stranded DNA Fragment Size Using the Wizard® SV Gel and PCR Clean-Up System.
| DNA Fragment Size | Percent Recovery |
|---|---|
| 55bp | 26% |
| 70bp | 39% |
| 85bp | 55% |
| 100bp | 84% |
| 500bp | 89% |
| 1,000bp | 92% |
| 3,199bp | 95% |
| 9,416bp | 95% |
| 23,130bp | 47% |
Table 8. Effect of Various PCR Additives on Percent Recovery of a 1,000bp PCR Product Using the Direct Purification Method and the Wizard® SV Gel and PCR Clean-Up System.
| PCR Additive | Percent Recovery1 |
|---|---|
| no additive | 100% |
| 1M betaine | 94% |
| 1M Q-Solution | 97% |
| 0.1% Triton® X-100 | 92% |
| 0.1% Tween®-20 | 87% |
| 0.1% NP-40 | 82% |
| 5% glycerol | 87% |
| 5% formamide | 90% |
| 5% DMSO | 87% |
| 0.5M tetramethylene sulfoxide | 94% |
| 0.4M sulfolane | 94% |
| 0.4M 2-pyrollidone | 95% |
| 1mM tartrazine | 100% |
| 1% Ficoll®-400 | 100% |
For direct purification from a reaction, note that any nucleic acid present in solution will be isolated. Therefore, if an amplification reaction has more than one product, all fragments will be present in the eluted DNA. If you are interested in isolating a single amplicon, separate the reaction products on an agarose gel and cut out the band desired prior to purification. When purifying DNA from an agarose slice, the primary consideration is to melt the agarose so the DNA is available for binding to the silica membrane. The purified DNA can then be used for cloning or sequencing.
The Wizard® and ReliaPrep™ clean-up kits have similar capabilities, however the ReliaPrep™ kit is better suited to performing more significant concentrations and can be completed in less time.
Wizard® SV 96 PCR Clean-Up System
To purify 96 amplification reactions at once, use the Wizard® SV 96 PCR Clean-Up System (Cat.# A9340, A9341, A9342, A9345) Wizard® SV 96 PCR Clean-Up System with a 96-well vacuum manifold (Vac-Man® 96 Vacuum Manifold) and a vacuum pump capable of generating 15–20 inches of mercury or the equivalent. This system is designed to purify 100bp to 10kb PCR products directly from a reaction with typical recovery >90% as seen in Figure 21.
The technology is the same as the single-column system, utilizing the SV silica membrane and chaotropic salts to purify the nucleotides and primers from the PCR product(s). This system allows recovery of 96 PCR fragments in as little as 20 minutes in multiwell plate format. The DNA can be used for automated fluorescent DNA sequencing, cloning, labeling, restriction enzyme digestion or DNA microarray analysis without further manipulation.
Figure 21. Purification and recovery of PCR products using the Wizard® SV 96 PCR Clean-Up System. PCR fragments of 100, 200, 300, 500 and 1,000 base pairs were purified using the Wizard® SV 96 PCR Clean-Up System on the Biomek® 2000 robotic workstation. Panel A. Agarose gel analysis. Purified (P) and unpurified (U) fragments were separated on an ethidium bromide-stained, 2% agarose gel. Panel B. Percent recovery of purified PCR products. Percent recovery was quantitated using a Hitachi FMBIO® Fluorescent Scanner. Results show the mean and standard deviation for 6 purified fragments of each size. For small PCR fragments (<500bp), optimal recovery requires a 95% ethanol wash. For larger fragments (>500bp), optimal results are achieved using an 80% ethanol wash.
Sequencing Cleanup
ProNex® Size-Selective Purification System
The ProNex® Size-Selective Purification System (Cat.# NG2001, NG2002, NG2003) enables the rapid and efficient magnetic resin-based purification of double-stranded DNA (dsDNA) for NGS, PCR and general molecular biology applications. The ProNex® System allows users to select the desired size of purified dsDNA fragments, from 100bp to 750bp. The novel reagent formulation provides significantly improved selectivity, reproducibility and yield relative to traditional dsDNA purification methods. In addition, the ProNex® System can be used in both manual and automated high-throughput workflows.
BigDye® Sequencing Cleanup
Designed for BigDye® Sanger sequencing reaction cleanup, the Wizard® MagneSil® Sequencing Reaction Clean-Up System (Cat.# A1831, A1832, A1835) can be placed on a robotic platform and purified using the MagneSil® PMPs to clean up sequencing reaction products prior to analysis. We have developed procedures for use on several robotic workstations with standard 96- and 384-well amplification plates. The Plate Clamp 96 (Cat.# V8251) is recommended for automated protocols and is designed to ensure PCR plates are uniformly flat for liquid transfer on a robotic platform. No user intervention is required from the time the multiwell plates are placed on the robot deck until the samples are loaded onto the DNA sequencer.
Methods for Determining DNA Yield & Purity by Absorbance
DNA yield can be assessed using three different physical methods: absorbance (optical density), agarose gel electrophoresis and fluorescent DNA-binding dyes. Each technique is described below and includes information on necessary accessories (e.g., equipment). While all methods are useful, each has caveats to consider when choosing a quantitation approach.
The most common technique to determine DNA yield and purity is also the easiest method—absorbance. All that is needed for measurement is a spectrophotometer equipped with a UV lamp, UV-transparent cuvettes (depending on the instrument) and a solution of purified DNA. Absorbance readings are performed at 260nm (A260) where DNA absorbs light most strongly, and the number generated allows one to estimate the concentration of the solution. To ensure the numbers are useful, the A260 reading should be between 0.1–1.0.
However, DNA is not the only molecule that can absorb UV light at 260nm. Since RNA also has a great absorbance at 260nm, and the aromatic amino acids present in protein absorb at 280nm, both contaminants, if present in the DNA solution, will contribute to the total measurement at 260nm. Additionally, the presence of guanidine will lead to higher 260nm absorbance. This means that if the A260 number is used for calculation of yield, the DNA quantity may be overestimated (43).
To evaluate DNA purity by spectrophotometry, measure absorbance from 230nm to 320nm in order to detect other possible contaminants present in the DNA solution. The most common purity calculation is determining the ratio of the absorbance at 260nm divided by the reading at 280nm. Good-quality DNA will have an A260/A280 ratio of 1.7–2.0. A reading of 1.6 does not render the DNA unsuitable for any application, but lower ratios indicate more contaminants are present. However, the best test of DNA quality is functionality in the application of interest (e.g., real-time PCR).
Strong absorbance around 230nm can indicate that organic compounds or chaotropic salts are present in the purified DNA. A ratio of 260nm to 230nm can help evaluate the level of salt carryover in the purified DNA. The lower the ratio, the greater the amount of thiocyanate salt is present, for example. As a guideline, the A260/A230 is best if greater than 1.5. A reading at 320nm will indicate if there is turbidity in the solution, another indication of possible contamination. Therefore, taking a spectrum of readings from 230nm to 320nm is most informative.
Agarose gel electrophoresis of the purified DNA eliminates some of the issues associated with absorbance readings. To use this method, a horizontal gel electrophoresis tank with an external power supply, analytical-grade agarose, an appropriate running buffer (e.g., 1X TAE) and an intercalating DNA dye along with appropriately sized DNA standards are needed for quantitation. A sample of the isolated DNA is loaded into a well of the agarose gel and then exposed to an electric field. The negatively charged DNA backbone migrates toward the anode. Since small DNA fragments migrate faster, the DNA is separated by size. The percentage of agarose in the gel will determine what size range of DNA will be resolved with the greatest clarity (40). Any RNA, nucleotides and protein in the sample migrate at different rates compared to the DNA so the band(s) containing the DNA will be distinct.
Concentration and yield can be determined after gel electrophoresis is completed by comparing the sample DNA intensity to that of a DNA quantitation standard. For example, if a 2µl sample of undiluted DNA loaded on the gel has the same approximate intensity as the 100ng standard, then the solution concentration is 50ng/µl (100ng divided by 2µl). Standards used for quantitation should be labeled as such and be the same size as the sample DNA being analyzed. In order to visualize the DNA in the agarose gel, staining with an intercalating dye such as ethidium bromide or SYBR® Green is required. Because ethidium bromide is a known mutagen, precautions need to be taken for its proper use and disposal (43).
DNA-binding dyes compare the unknown sample to a standard curve of DNA, but genomic, fragment and plasmid DNA will each require their own standard curves and cannot be used interchangeably. If the DNA sample has been diluted, you will need to account for the dilution factor when calculating final concentration. Hoechst bisbenzimidazole dyes or PicoGreen® selectively bind double-stranded DNA (dsDNA). To use this method, a fluorometer to detect the dyes, dilution of the DNA solution and appropriate DNA standards are required. However, there are size qualifications: the DNA needs to be at least 1 kilobase in length for Hoechst and at least 200bp for PicoGreen® for successful quantitation. The range of measurement is 10–250ng/ml for Hoechst, 25pg/ml–1µg/ml for PicoGreen®, and the dyes are sensitive to GC content. In addition, the usual caveats for handling fluorescent compounds apply—photobleaching and quenching will affect the signal. While the dyes bind preferentially to dsDNA, RNA and nucleotides may contribute to the signal.
Choosing which quantitation method to use is based on many factors including access to equipment or reagents, reliability and consistency of the concentration calculations. Use caution when comparing yields between methods as the level of potential contaminants may cause variable determinations among the different methods.
Webinar: To NanoDrop or Not to NanoDrop: Choosing the Most Appropriate Method for Nucleic Acid Quantitation
Learn about the advantages and disadvantages of current DNA/RNA quantitation methods, including absorbance, fluorescent nucleic acid-binding dyes and qPCR.
How to Determine the Concentration, Yield and Purity of a DNA Sample
This article explains the various methods for determining DNA yield.
Summary
In this DNA purification guide, we discussed the basic steps of DNA extraction, plasmid preparation and DNA quantitation, and explored the vast portfolio of products that Promega has to offer.
This guide is intended to help you understand those basics, navigate issues of scalability, purity, yield and the effects they have on downstream applications, and ultimately assist you in identifying the system that best fits your DNA purification needs.
Need additional assistance? Here at Promega, your success is important to us and we genuinely enjoy the challenge of identifying the right product to address your technical needs. Our Technical Services department is available to help guide you every step of the way, from answering technical questions about your products to providing support for your automated instruments.
Related Products
References
- Mandrekar, P. (2016) Introduction to Nucleic Acid Purification: Purification Basics and Their Application to Different Sample Types [Webinar].
- Chen, C.W. and Thomas, C.A. Jr. (1980) Recovery of DNA segments from agarose gels. Anal. Biochem.101, 339–41.
- Marko, M.A. et al. (1982) A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal. Biochem. 121, 382–7.
- Boom, R. et al. (1990) Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28, 495–503.
- Melzak, K.A. et al. (1996) Driving forces for DNA adsorption to silica in perchlorate solutions. J. Colloid Interface Sci. (USA)181, 635–44.
- Walker, J.A. et al. (2003) Quantitative intra-short interspersed element PCR for species-specific DNA identification. Anal. Biochem.316, 259–69.
- Zhang, X. et al. (2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 39, 905–19.
- van Schaik, W. et al. (2004) The alternative sigma factor σB of Bacillus cereus: Response to stress and role in heat adaptation. J. Bacteriol.186, 316–25.
- Flashner, Y. et al. (2004) Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun. 72, 908–15.
- Lee, P.H. et al. (2005) Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc. Natl. Acad. Sci. USA.102, 3750–55.
- Martinez, C. et al. (2004) Genomic characterization of Saccharomyces cerevisiae strains isolated from wine-producing areas in South America. J. Appl. Microbiol.96, 1161–8.
- Ahmed, A. et al. (2003) Madurella mycetomatis strains from mycetoma lesions in Sudanese patients are clonal. J. Clin. Microbiol. 41, 4537–41.
- Pereira, C.N. et al. (2005) The pathogen of frogs Amphibiocystidium ranae is a member of the order dermocystida in the class mesomycetozoea. J. Clin. Microbiol.43, 192–8.
- Cox, A.J. et al. (2004) Valtrex therapy for Epstein-Barr virus reactivation and upper respiratory symptoms in elite runners. Med. Sci. Sports Exerc. 36, 1104–10.
- Lorenzen, M.D. et al. (2002) Cloning and characterization of the Tribolium cataneum eye-color genes encoding tryptophan oxygenase and kynurenine 3-monooxygenase. Genetics. 160, 225–34.
- Promega Corporation (2002) Genomic DNA purification from blood. Application Notes. 101, 1–4.
- Stump, A.D. et al. (2005) Sex-linked differentiation between incipient species of Anopheles gambiae. Genetics. 169, 1509–19.
- Dontu, G. et al. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev.17, 1253–70.
- Park, J.H. et al. (2004) Characterization of two kinases involved in thiamine pyrophosphate and pyridoxal phosphate biosynthesis in Bacillus subtilis: 4-amino-5-hydroxymethyl-2methylpyrimidine kinase and pyridoxal kinase. J. Bacteriol.186, 1571–3.
- Teresa Pellicer, M. et al. (2003) Role of 2-phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J. Bacteriol.185, 5815–21.
- Smith, P. et al. (2004) Schistosoma mansoni worms induce anergy of T cells via selective up-regulation of programmed death ligand 1 on macrophages. J. Immunol.173, 1240–8.
- Ohsaki, E. et al. (2004) Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol.78, 9936–46.
- Birnboim, H.C. and Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucl. Acids Res.7, 1513–23.
- Birnboim, H.C. (1983) A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol.100, 243–55.
- Hirt, B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol.26, 365–9.
- Holmes, D.S. and Quigley, M. (1981) A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem.114, 193–7.
- Hamaguchi, K. and Geiduschek, E.P. (1962) The effect of electrolytes on the stability of the deoxyribonucleate helix. J. Am. Chem. Soc. 84, 1329–38.
- Wilcockson, J. (1973) The use of sodium perchlorate in deproteinization during the preparation of nucleic acids. Biochem. J.135, 559–61.
- Wilcockson, J. (1975) The differential precipitation of nucleic acids and proteins from aqueous solutions by ethanol. Anal. Biochem. 66, 64–8.
- van Huynh, N. et al. (1993) Sequential elution of denatured proteins, hydrolyzed RNA, and plasmid DNA of bacterial lysates adsorbed onto stacked DEAE-cellulose membranes. Anal. Biochem. 211, 61–5.
- Lis, J.T. (1980) Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods Enzymol.65, 347–53.
- Paithankar, K.R. and Prasad, K.S. (1991) Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 19, 1346.
- Wang, Z. and Rossman, T.G. (1994) Isolation of DNA fragments from agarose gel by centrifugation. Nucl. Acids Res.22, 2862–3.
- 34. Ausubel, F.M. et al. (1989) Current Protocols in Molecular Biology, Vol. 2, John Wiley and Sons, NY.
- Davies, J. and Smith, D.I. (1978) Plasmid-determined resistance to antimicrobial agents. Annu. Rev. Microbiol. 32, 469–518.
- Lehman, I.R. et al. (1962) The deoxyribonucleases of Escherichia coli. II. Purification and properties of a ribonucleic acid-inhibitable endonuclease. J. Biol. Chem.237, 819–28.
- Goebel, W. and Helinski, D.R. (1970) Nicking activity of an endonuclease. I. Transfer ribonucleic acid complex of Escherichia coli. Biochemistry. 9, 4793–801.
- Shortman, K. and Lehman, I.R. (1964) The deoxyribonucleases of Escherichia coli. VI. Changes in enzyme levels in response to alterations in physiological state. J. Biol. Chem.239, 2964–74.
- Schoenfeld, T. et al. (1995) Effects of bacterial strains carrying the endA1 genotype on DNA quality isolated with Wizard® Plasmid Purification Systems. Promega Notes. 53, 12–20.
- Sambrook, J. et al. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edition Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Lewin, B. (2004) In: Genes VIII Pearson Prentice Hall, Upper Saddle River, NJ, 379–80.
- Butash, K.A. et al. (2000) Reexamination of the effect of endotoxin on cell proliferation and transfection efficiency. BioTechniques. 29, 610–9.
- 43. Adams, D.S. (2003) In: Lab Math: A Handbook of Measurements, Calculations, and Other Quantitative Skills for Use at the Bench Chapter 5, Cold Spring Harbor Laboratory Press, NY, 127–45.