Targeting RNA
Small molecules and oligos that target RNA or RNA-modifying proteins are emerging as a promising therapeutic approach. These molecules can modulate RNA function in a variety of ways, such as changing expression levels, promoting exon skipping and influencing epitranscriptomics.
Understanding the function of various RNAs and the proteins they interact with can be challenging. Further, determining the specificity and selectivity of small molecules and oligos requires sensitive assays. Promega offers a comprehensive selection of technologies to enable your RNA research and help answer these important questions.
Share your research goals with us in the area of RNA biology, and let our team find a custom solution for you!
Structural model of risdiplam, the first small-molecule drug targeting RNA that was approved by the US Food and Drug Administration (FDA). The drug is being used to treat spinal muscular atrophy. See Sheridan, C. (2021) Nature Biotechnol. 393–12. DOI: 10.1038/s41587-020-00788-1.
Though RNA was once considered a passive messenger, we now appreciate its complexity and understand its implications in disease. There are many different types of cellular RNAs, and much remains to be understood, including their structure and function. More than 1,500 proteins bind RNA; yet there is much to learn on what these proteins do and how they do it. Oligonucleotides—such as small interfering RNA (siRNA), antisense, aptamers and other RNA moieties—have traditionally been used to target RNA. Now, small molecules have joined these modalities to target the three-dimensional (3D) structure of RNA directly. Much progress has been made in binding and inhibiting RNA, and the field continues to investigate challenges such as specificity and cell permeability.
Expression Level
Monitor dose- and time-dependency of changes in expression using CRISPR-HiBiT cell lines

Specificity and rank ordering of siRNAs. Kinetic monitoring of endogenous cMyc was performed in the cMyc-HiBiT KI HEK293 (LgBiT) cells, which stably express LgBiT, allowing kinetic studies. Cells were treated with siRNAs against cMyc or GAPDH. Nano-Glo® Endurazine™ Live Cell Substrate was added at the time of transfection, and luminescence was monitored every 10 minutes on a GloMax® Discover Microplate Reader.
Multiplex cell viability assays with your expression analysis to determine if there is an effect on cell health

Determine cytotoxicity of your treatment by multiplexing cell health assays in the same wells as your expression analysis. cMyc-HiBiT KI HEK293 (LgBiT) cells were treated with siRNAs against cMyc or a non-targeting siRNA (Dharmacon) for 48 hours. First, cell viability was measured using the CellTiter-Fluor™ Cell Viability Assay normalized to untransfected cells. Next, the expression of the cMyc:HiBiT fusion was measured using the Nano-Glo® HiBiT Lytic Detection System. Bars show cMyc:HiBiT (%) relative to the negative control siRNA. Dots show cell viability relative to the untransfected controls.
Monitor expression through dual-reporter systems
As a world leader in bioluminescent reporter technology, we have a portfolio of dual-luciferase reporter assays that enable fast and simple quantitation of a stable luminescent signal from two reporter genes in a single sample. The activity of the primary reporter is correlated with the effect of specific stimuli, and the activity of the co-transfected control reporter provides an internal control to normalize results.
You can use the Dual-Glo® Luciferase Assay System for the classic firefly and Renilla luciferase combination. If you need more sensitivity (e.g., low cell number, challenging cell line, etc.), take advantage of the substantial increase in brightness that NanoLuc® luciferase provides, or consider the state-of-the-art Nano-Glo® Dual-Luciferase® Reporter Assay System.
Analyze endogenous microRNA, mi-R21, in HeLa cells by normalizing luciferase activity
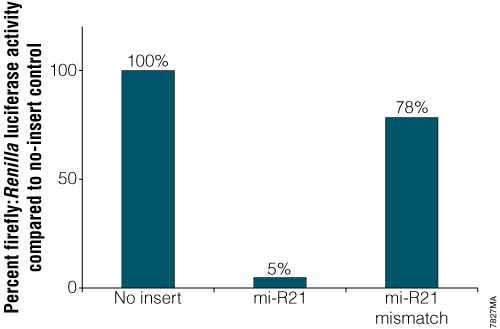
Analysis of endogenous miRNA. Constructs contained either an exact match to the 21bp mi-R21 target sequence or a mismatched version of that target site. Twenty-four hours after transfection with the mi-R21 pmirGLO Vector constructs, cells were analyzed for luciferase activity using the Dual-Glo® Luciferase Assay System. Normalized firefly luciferase activity (firefly luciferase activity/Renilla luciferase activity) for each construct was compared to that of the pmirGLO Vector no-insert control.
Monitor expression at the nucleic acid level with manual, automated and high-throughput options
Maxwell® RSC technology includes two compact, automated nucleic acid purification instruments that process up to 16 or 48 samples simultaneously. These magnetic particle movers use prefilled cartridges and preprogrammed methods to extract DNA or RNA from a wide variety of sample types in 25–60 minutes. Learn how Maxwell® instruments enable you to extract high-quality nucleic acids with less hands-on time.
Need larger scale extractions? Our Maxwell® chemistries can be freed from the cartridges and used with a wide range of high-throughput liquid handling platforms. Bonus: we have a team of experts who have extensive experience with liquid handling platforms. They will set up our chemistries on your liquid handler for you. Learn more about our high-throughput extraction chemistries and our expert liquid handling team.
Working on a smaller scale and don’t need automation yet? We can help with manual extraction solutions for RNA and DNA, too.
See our Application Notes for manual and automated DNA or RNA extraction protocols from various sample types.
Automated nucleic acid extraction with Maxwell® technology followed by RT-qPCR

Get more data from the same well by combining our cell viability assays with nucleic acid extraction. Read this application note to see how we measured cell viability in real time and then monitored gene expression from the same well during siRNA gene silencing.
RNA-Protein Interactions
The interplay between RNA and RNA-binding proteins (RBPs) is complex, with RNA often bound and modified by RBPs. There are over 1,500 RBPs, and we have much to learn about their function and binding specificities. Disruption of these interactions is implicated in many diseases, from neurodegeneration to cardiovascular disease to cancer.
Please contact us to discuss terms and conditions for applications using labeled nucleotides as energy transfer acceptors.
Use NanoBRET® technology to measure binding affinities of oligos to your protein of interest

Schematic overview of a NanoBRET® binding affinity assay. Fuse NanoLuc® luciferase to your target protein of interest and express in cells. Label an RNA tracer oligo with a fluorophore, and allow binding to the fusion protein. Lyse cells with digitonin, and measure binding affinities of different unlabeled RNA oligos by competitive displacement of the tracer oligo.
Use NanoBiT® and HaloTag® technologies to analyze RNA-protein interactions
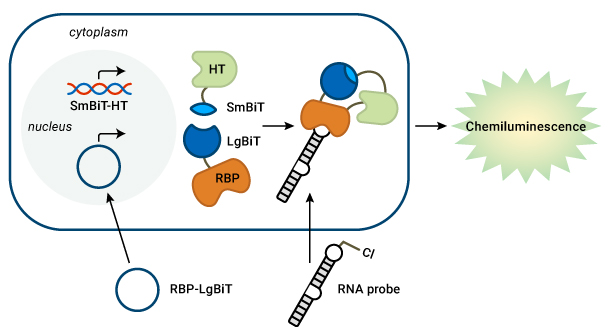
Detect RNA-RBP interactions in live cells. Prof. Garner’s lab transfects an RBP-LgBiT plasmid and a chloroalkane-labeled pre-miRNA probe into cells that stably express SmBiT-HaloTag (HT). The HT covalently binds to the RNA probe. When the RNA and RBP interact, the BiTs can assemble to form a functional luciferase enzyme and generate light.
For more information on the RiPCA method, read our blog interview with Prof. Garner, or see the publication in RSC Chemical Biology.
Epitranscriptomics
Detect RNA demethylation using the Succinate-Glo™ Assay

Measure RNA demethylase activity in a sensitive bioluminescent assay
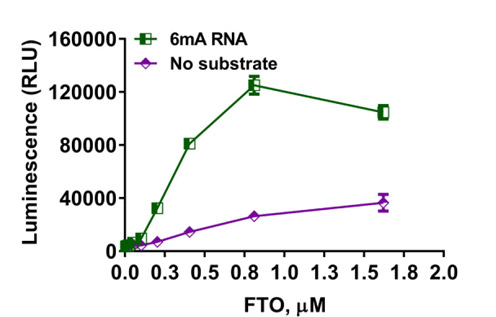
Determination of demethylase activity. The Succinate-Glo™ Assay was used to detect the activity of RNA demethylase FTO. FTO was titrated in the presence or absence of 10µM of 6mA-RNA.
Measure RNA methyltransferase activity
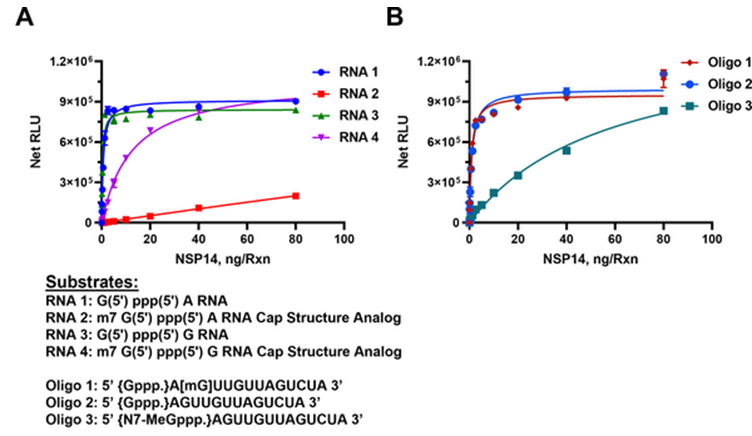
Monitor the activity of RNA methyltransferases (MTases) by detecting the formation of the reaction product S-adenosyl homocysteine (SAH). Learn more about the assay on the product page and in this publication.
The MTase-Glo™ Assay was used to determine SARS-CoV-2 nsp14 MTase activity using capped and uncapped RNA substrates as listed under the graphs. Panel A. Short sequence substrates. Panel B. Long sequence substrates. Learn more about the study in this publication.
siRNA and miRNA
Are you modulating RNAs using siRNA or miRNAs? We have vectors ready to go that will allow you to monitor siRNA and miRNA activity using dual-luciferase technology.
- The pmirGLO Dual-Luciferase Vector allows you to insert miRNA target sites 3´ of the firefly luciferase gene to monitor activity. Plus it has an internal Renilla luciferase control for normalization.
- The psiCHECK™ Vector allows you to monitor siRNA activity through degradation of a Renilla luciferase. Clone your region of interest downstream of the Renilla luciferase gene, and initiation of RNAi will result in cleavage and degradation of the mRNA fusion. An internal firefly luciferase control allows for easy normalization.
Need a way to introduce siRNAs or miRNAs into your eukaryotic cell line? The FuGENE® SI Transfection Reagent is designed specifically for the delivery of small RNA molecules while minimizing impact on cell health.
Need to produce duplex RNA for your studies? The T7 RiboMAX™ Express RNAi System produces milligram amounts of RNA, both siRNA duplexes for mammalian RNAi and long dsRNA for nonmammalian RNAi.
shRNA optimization for RNAi of bcr/abl
A 200bp region of the bcr/abl mRNA that spans the junction region between the bcr and abl genes of the hybrid mRNA was amplified by RT-PCR from K562 cells, a human CML cell line that is positive for the Philadelphia chromosomal translocation (GenBank® accession# M30829) and cloned into the psiCHECK™-2 Vector.
Seven shRNAs to the bcr/abl mRNA were synthesized using the T7 RiboMAX™ Express RNAi System. Chinese hamster ovary cells (CHO) were transfected with the psiCHECK™-2-bcr/abl plasmid as well as the various shRNAs. The Dual-Glo™ Luciferase Assay System was used to measure Renilla luciferase and firefly luciferase reporter activity.
Read the full study.

RNA Analysis


Gel analysis of RNA integrity after incubation. Five microliters of each reaction were mixed with 4µl of formaldehyde sample buffer (Lonza Cat.# 50571) and 2µl of 0.1mg/ml ethidium bromide. The mixtures were incubated at 65°C for 5 minutes. Six microliters of each mixture were run on a 1% agarose gel with 1X TBE buffer at 100V for approximately 25 minutes.
Read more information in the technical article.
Learn about our entire RNA analysis portfolio, which also includes:
- fluorescent dyes for RNA quantification
- in vitro transcription reagents (e.g., large-scale RNA production systems (RiboMAX), and systems for generation of RNA probes (Riboprobe))
- RT-PCR enzymes and kits (e.g., standalone reverse transcriptases (AMV, M-MLV and GoScript) and a range of complete kits containing all the required enzymes, buffers and reagents for successful RT-PCR)
Profiling Services
Our custom assay services can help to accelerate your drug discovery and development workflow. These comprehensive services are built around our technologies for sensitive, high-throughput and biologically relevant results. For more information, contact the Tailored R&D Solutions team.
- CRISPR-HiBiT cell-line screening
- Live-cell target engagement
- RNA-protein interactions
- Reporter gene assays
- And more