Reproducible Drug Screening Assays Using Single Organoids
Cell Microsystems and Promega Corporation
Publication Date: May 2023
Introduction
Three-dimensional (3D) cell culture models are at the forefront of transforming drug discovery and personalized medicine. By more faithfully recapitulating the pathophysiology of human tissues in vitro, these models have the potential to fill the “valley of death” translation gap between 2D cell-based assays and human responses. Organoids, which can be derived from both healthy and diseased adult stem cells, including tumor cells or induced pluripotent stem cells (iPSCs), offer a unique advantage over other 3D models, because they more accurately represent the cellular heterogeneity of tissues and maintain genomic and phenotypic profiles of the source material. Despite the pressing need for such models in pharmacology and toxicology, adoption of organoid in vitro assays for high-throughput screening is still limited due to workflow challenges, including lack of reproducibility and scalability, which impacts downstream assessment.
Together, CellRaft® Technology from Cell Microsystems and Promega Cell Health Assays are addressing two critical bottlenecks preventing the acceleration of organoid models into high-throughput screening applications by providing:
- an automated solution for generating customized, scalable and reproducible organoids that are ready to use in plate-based assays; and
- assays that have been optimized and validated for 3D cell culture models, including those that enable real-time continuous measurements.
Creating Custom Organoid Assays Using CellRaft® Technology
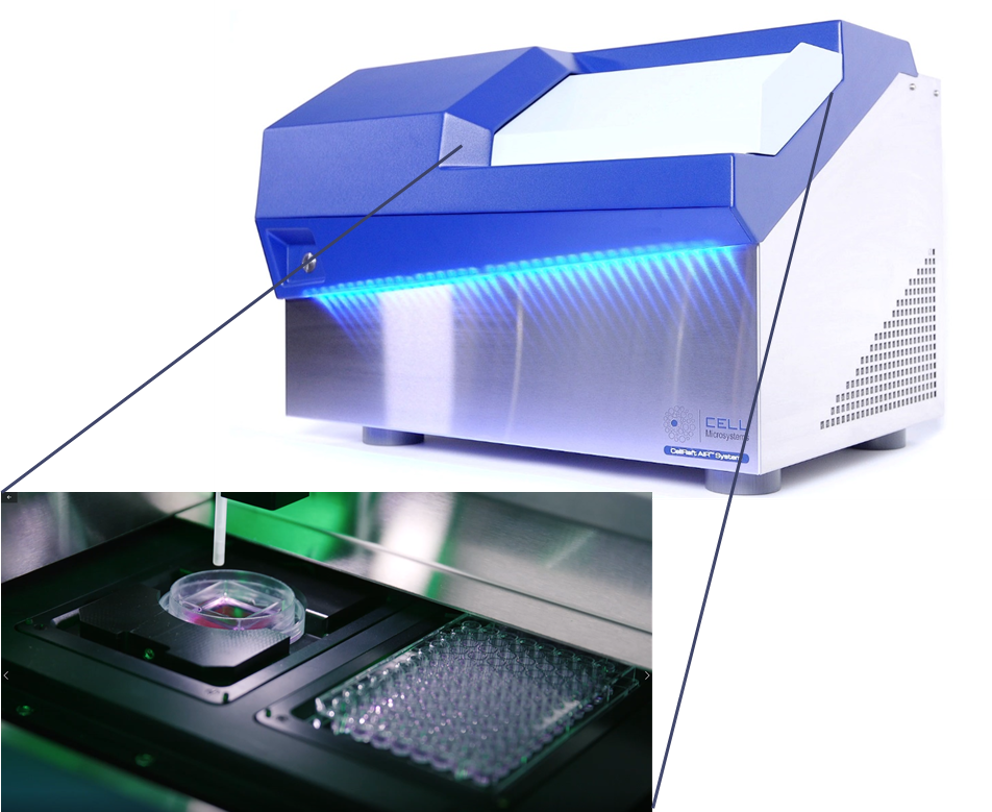
Improving the scalability, efficiency and reproducibility of organoid models for screening applications is one of the biggest challenges researchers face in using these complex 3D models for their research and discovery programs. CellRaft® Technology offers a unique solution to many of the pain points researchers face in their organoid workflows. Using the core technology, the CellRaft® Array and the CellRaft® AIR® System (Figure 1), users can:
- Grow and maintain hundreds of spatially segregated organoids in a single focal plane on the CellRaft® Array using less media and reagents.
- Perform automated imaging to capture serial images of each organoid over time.
- Use software tools to phenotypically characterize the heterogeneous population of organoids grown on the CellRaft® Array and identify organoids of interest, based on phenotypic and morphologic properties.
- Isolate single, intact organoids for downstream use, including drug screening assays.
A.
B.
C.
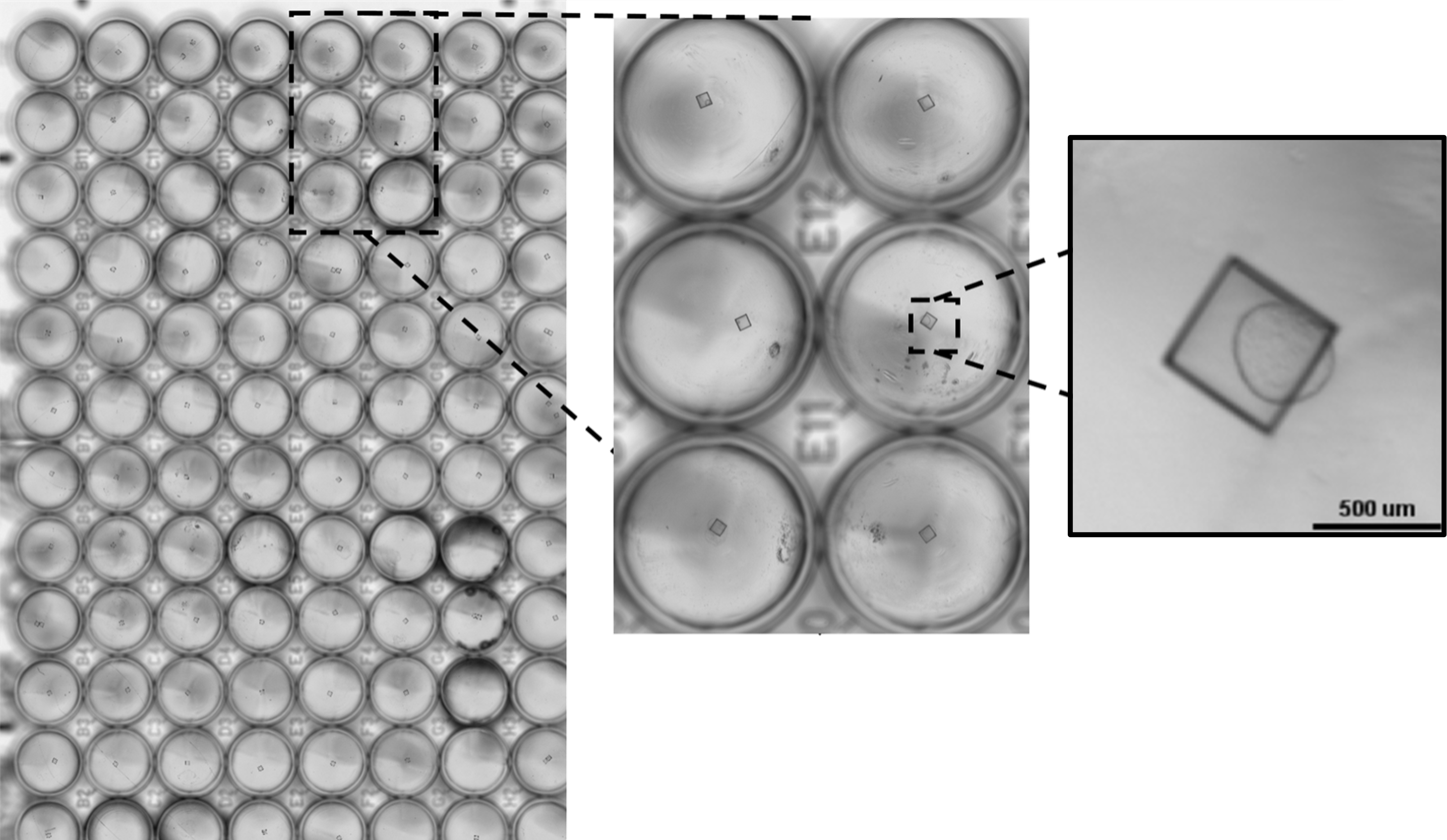
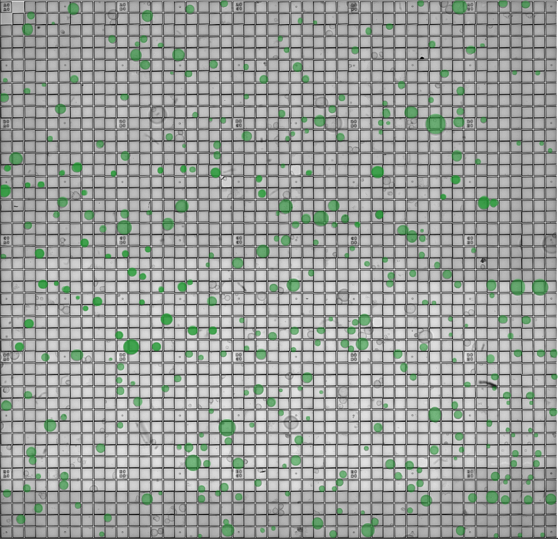
Figure 1. The CellRaft® Array and CellRaft® AIR System enable automated imaging, identification and isolation of hundreds of individual organoids. Field-of-view images, acquired on the CellRaft® AIR System, were stitched together to generate an image of the entire CellRaft® Array (A). Green circles highlight every CellRaft® in the array containing a single organoid. Each CellRaft® can be reliably imaged over time to identify CellRafts containing single organoids of interest for downstream assays. Using the CellRaft® AIR System (B) CellRafts, with attached organoids of interest, were isolated into 96-well collection plates to generate customized organoid assay plates with a single organoid in each well (C).
Promega Plate-Based Cell Health Assays Optimized and Validated for 3D Cell Culture Models
Cell health assays from Promega have been routinely implemented with different 3D cell culture models and, where necessary, were specifically optimized with enhanced reagents (e.g., CellTiter Glo® 3D Cell Viability Assay) or use modified protocols (e.g., Caspase Glo® 3/7 3D Assay) for higher capacity to penetrate 3D tissues. Details for each assay used in this article can be found in Figure 2.
A.
B.

C.
D.
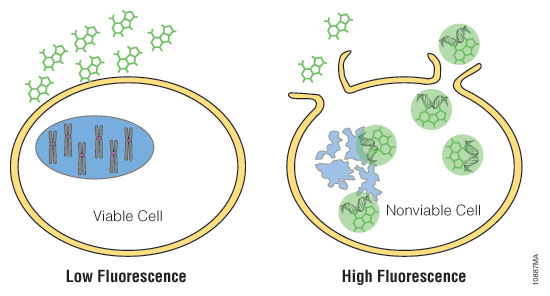
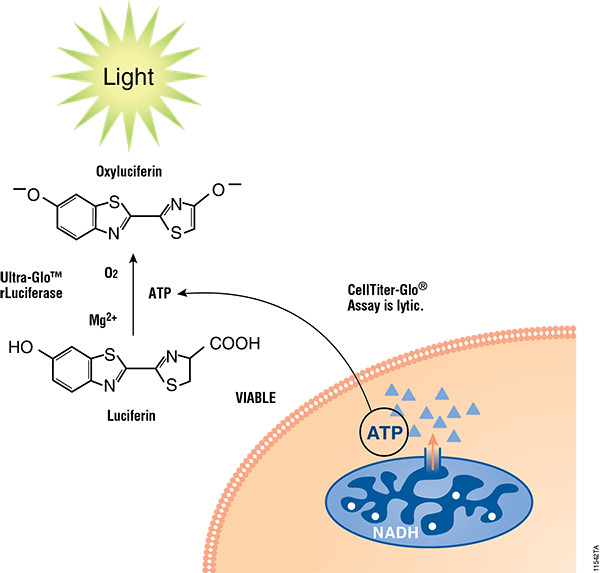
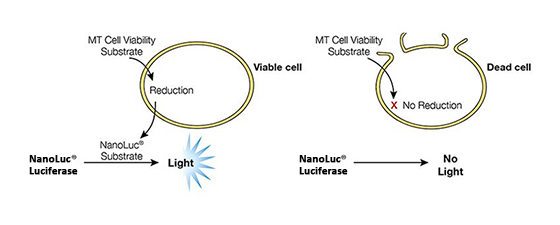
Figure 2. Mechanisms of assay panel. CellTiter-Glo® 3D Cell Viability Assay (A) is a lytic reagent that quantifies ATP content in a firefly luciferase reaction. Luminescence is proportional to viable cell number. Caspase Glo® 3/7 3D Assay (B) is a lytic reagent comprised of a Z-DEVD pro-luciferin substrate that is cleaved by caspase-3/7 to produce luciferin. Luminescence is proportional to caspase-3/7 activity. In the RealTime-Glo™ MT Cell Viability Assay (C), a nonlytic NanoLuc® Luciferase reaction occurs with substrate that has been reduced by viable cells. Luminescence is proportional to viable cell number. CellTox™ Green Cytotoxicity Assay (D) is a nonlytic reagent that binds to the DNA of nonviable cells. Fluorescence is proportional to dead cell number.
Case Study #1: Organoid Size Impacts Drug Screening Assays
One limitation to using conventional bulk cultures of organoids for drug and toxicity screening is the variability of organoid number and size between wells in an experiment and across multiple experiments. We used the CellRaft® Array and CellRaft® AIR System to culture, image and isolate mouse hepatic organoids to demonstrate the ability to create consistent, reproducible organoid assays for toxicity testing. Using CellRaft® Cytometry, CellRafts containing a single mouse hepatic organoid were selected for isolation based on organoid diameter. Two distinct populations were isolated using the CellRaft® AIR System to evaluate assay consistency. The first population consisted of variably sized organoids (>50µm), and the second population was size-selected to identify organoids that were 300–500µm in diameter. We used two approaches to demonstrate the importance of organoid size on assay consistency. Kinetic viability of untreated organoids from the unselected and size-selected populations (n = 50 wells per population) was measured every 24 hours for 72 hours using the RealTime-Glo™ MT Cell Viability Assay. In addition, single organoids from each population were treated in parallel with a six-point, five-fold dose curve of acetaminophen (APAP; 0.0008–2.5mM) with five replicate wells per dose. Toxicity assessment was performed using two Promega cell heath assays. Kinetic viability was measured every 24 hours for 72 hours using the CellTox™ Green Cytotoxicity Assay, and total ATP was measured after 72 hours of treatment using the CellTiter-Glo® 3D Cell Viability Assay.
Using CellRaft® Technology, we demonstrated that organoid size is critical to maintaining intra-assay consistency. As expected, kinetic viability of untreated organoids from the unselected size population demonstrated more well-to-well variability, compared to size-selected organoids (Figure 3). In addition, in the parallel toxicity dose curve, the size-selected replicates have smaller standard error values in both viability and ATP readouts (Figure 4B) compared to the unselected population of organoids (Figure 4A). Importantly, the well-to-well consistency of size-selected organoids was necessary for identifying an ED 50% (0.6mM), which was not possible in the dose curve using variable-sized organoids, due to the variance. These data demonstrate the ability to use CellRaft® Technology and software analysis tools for developing more consistent, reliable organoid screening assays for drug efficacy and toxicity within and between replicate experiments.

Figure 3. Kinetic viability of single mouse hepatic organoids isolated from the CellRaft® Array. Over time, organoids not selected for size (>50µm organoids) have greater well-to-well variability, compared to size-selected organoids (300–500µm organoids). Values for each group are reported as the mean (n = 50 organoids) with 95% confidence interval.
A.

B.

Figure 4. Dose-response curves for mouse hepatic organoids. Organoids were treated with a six-point dose curve of acetaminophen (APAP; 0.0008–2.5mM) with five replicate wells of each treatment for 72 hours. The left y-axis shows 24- (black), 48- (blue), and 72-hour (green) CellTox™ Green activity relative to the average DMSO control wells. The right axis shows the percent CellTiter-Glo® 3D activity relative to the DMSO control wells after 72 hours of treatment.
Case Study #2: Clonal iPSC-Derived Organoids for Drug Screening
Organoids derived from iPSCs have filled the gap in 3D models for tissues that are inaccessible or for organoids that cannot be developed from adult stem cells. In the last decade, protocols and commercial kits have been developed to differentiate iPSCs into multicellular, neuronal organoids that closely resemble human brain development, including region-specific cellular composition and functional physiology. These models are critical for modeling developmental and neurodegenerative disorders but have somewhat limited utility due to challenges of reproducibility and scalability. Using standard methodologies, bulk iPSCs are seeded into 96 well plates, pipetted into larger-well plates at multiple stages and manually embedded into Matrigel® droplets. In addition to being technically challenging, the standard workflow is limited to generating 96 organoids that cannot be tracked or analyzed throughout differentiation.
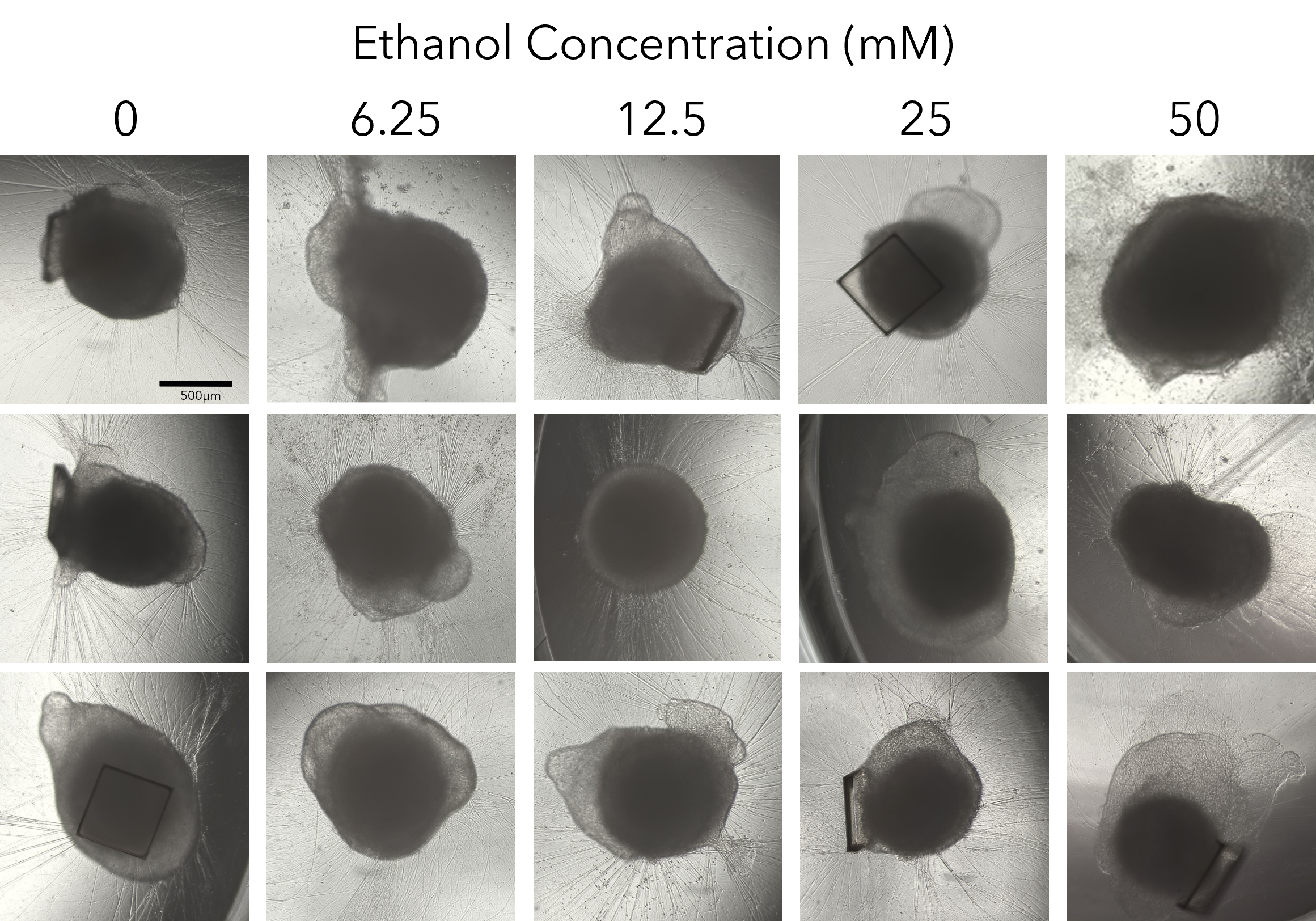
We collapsed this complex workflow onto the CellRaft® Array, providing a user-friendly method for growing hundreds of single iPSC-derived neuronal organoids for downstream toxicity screening. Single iPSCs were seeded and differentiated, through embryoid body formation, neural induction and expansion on the CellRaft® Array (Figure 5). On day 10, single cerebral organoids of interest, selected using CellRaft® Cytometry based on diameter (100–300µm), were isolated into maturation media using the CellRaft® AIR System. This process preserves organoid morphology and viability, and organoids were subsequently cultured off-array in cerebral maturation media out to day 42 (Figure 5). Images were captured of each organoid on day 40, after the cerebral organoids were fully mature. By selecting organoids for isolation based on day 10 size, we were able to generate a 96-well plate with single mature organoids that were relatively similar in size for our downstream toxicity screen (Figure 6A). On day 43, the organoids were treated with a six-point dose curve of ethanol (6.25–50mM) for 6 hours to simulate acute alcohol exposure. To evaluate alcohol-induced apoptosis, relative caspase activity was measured using the Caspase-Glo® 3/7 3D Assay. At the highest doses, we observed a dose-dependent activation of caspase activity, indicating alcohol-induced initiation of apoptosis (Figure 6B).
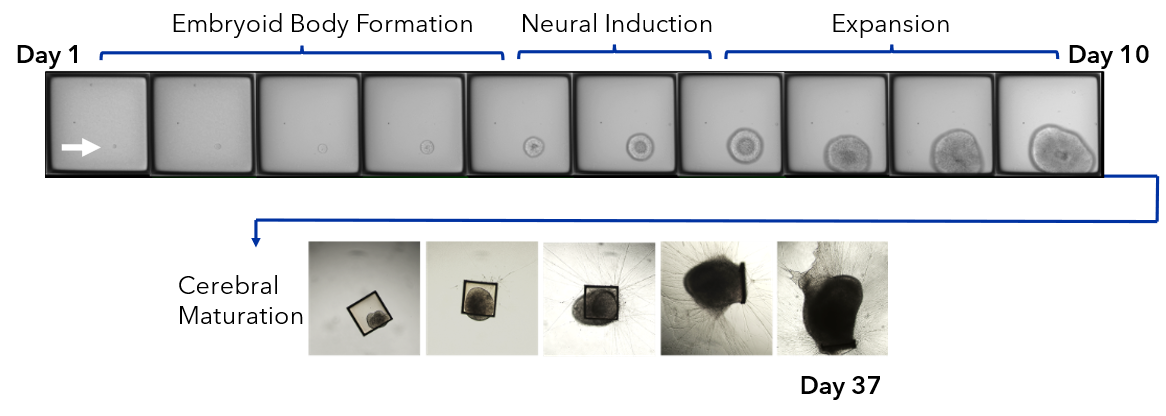
Figure 5. Single iPSCs are differentiated on the 500µm CellRaft® Array to form single neural organoids. iPSCs were seeded on the CellRaft® Array in dilute extracellular matrix (ECM) and imaged daily for a week during embryoid body formation and neural induction for choroid plexus organoid differentiation. On day 10, single organoids were selected based on diameter (100–300µm) and isolated into 96-well plates with maturation media. Cerebral organoids were maintained in maturation media to day 42.
A.
B.

Figure 6. Alcohol-induced apoptosis in neuronal organoids. Images of mature cerebral organoids on day 40, prior to treatment with ethanol, from three of the six replicate wells, demonstrating the ability to generate well-to-well reproducibility in the downstream assay on day 40 by selecting organoids based on size at day 10 (A). On day 43, mature cerebral organoids were exposed to a five-point dose curve of ethanol (6.25–50mM, n = 6 wells per dose) to simulate acute alcohol exposure. After 6 hours of treatment, organoids were evaluated for alcohol-induced apoptosis using the Caspase-Glo® 3/7 3D Assay. We observed a dose-dependent increase in caspase activity, supporting activation of apoptosis at the highest doses of ethanol exposure (B).
Conclusion
The biological power of 3D cell culture models is undermined by the technical challenges that scientists face attempting to leverage them for high-throughput plate-based screening assays. For these assays to gain wide adoption in screening, there is a need for technology that can enable the generation of organoids rapidly, reproducibly and cost-effectively. Using a single CellRaft® Array, hundreds of organoids can be grown at scale and monitored over time, using less media and ECM reagents compared to standard culture methodologies. Increasing organoid number, while reducing reagent usage, allows researchers to decrease the materials and labor burden of generating organoids, opening the door for high-throughput downstream assays. Using image-based, software-guided selection to isolate organoids based on size and other phenotypic characteristics improves well-to-well reproducibility for downstream assays and allows for normalization across multiple arrays and experiments. Lastly, the CellRaft® AIR System provides an automated solution for creating custom, 96-well plates of precharacterized organoids for downstream organoid screening assays. Together, CellRaft® Technology and Promega assay kits, which have been specifically designed and validated for 3D models, offer full spectrum, reliable organoid assay development for drug and toxicity screening.
Related Resources
3D Culture & Assays Webinar
In this webinar, our experts answer common questions about 3D culture and assays.
Evaluating Immunotherapy in Organoids
This blog post discusses how patient-derived colorectal organoids are used to evaluate CAR-NK immunotherapy.
3D Cell Culture Assays
An overview of assays and tools to monitor biology in 3D cell cultures.