Autophagy Detection
Autophagy is an important intracellular pathway for degradation and recycling of superfluous components, and for the removal of harmful subcellular materials. We offer an easy-to-use, plate-based autophagy detection assay that quantifies autophagic flux with a simple, bioluminescent signal.
The Autophagy LC3 HiBiT Reporter Assay discerns autophagy inducers from inhibitors with a quantitative, plate-reader based readout. This new autophagy detection method uses two reporter-based technologies: HiBiT and HaloTag, enabling plate-reader assays, protein blotting and imaging using a single reporter cell line. The versatile autophagy reporter allows quantitative study of the autophagy pathway, identification of novel modulators of autophagic flux, and confirmation of mechanism of action.
Filter By
Shop all Autophagy Detection
Showing 1 of 1 Products
Detecting Autophagic Flux
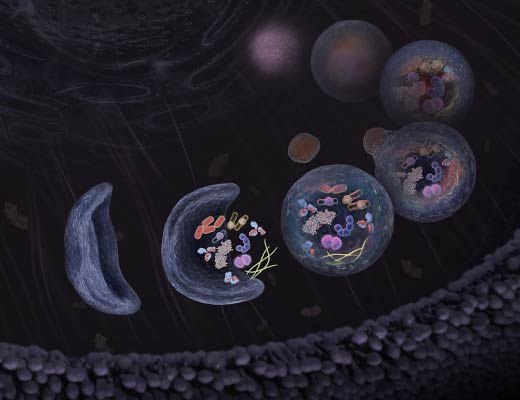
Autophagy is an intracellular degradation process that is implicated in many physiological and pathological conditions. When autophagy is induced, an isolation membrane is formed that encloses part of the cytoplasm and forms a double-membraned vesicle (autophagosome). The autophagosome outer membrane fuses with a lysosome to form the autolysosome, degrading the engulfed organelle components along with the inner autophagosome membrane. The term “autophagic flux” refers to the entire dynamic process of autophagy.
Mammalian LC3B is an autophagosome marker that is processed by Atg4 at its C-terminus to become LC3-I. LC3-I is mostly cytosolic, but is lipidated through phosphatidylethanolamine (PE) conjugation to form LC3-II that localizes at both the outer and inner membranes of the autophagosome. Upon autophagosome fusion with the lysosome, the inner-membrane bound LC3-II is degraded by lysosomal proteases. Monitoring the conversion of LC3-I to LC3-II by change in molecular weight is a common method to monitor changes in the autophagic pathway. Imaging LC3B localization in the cell from diffuse (in the form of cytostolic LC3-I) to concentrated (LC3-II in autophagosomes or puncta) is an additional method to track changes in autophagy.
There are several methods for monitoring autophagosome formation and autophagic flux. However, each method has its limitations, and more than one method should be used to validate autophagic activity. For example, using the number of autophagosomes determined by GFP-LC3 localization is a common method for measuring autophagy activity. However, the results might be misleading because both induction and inhibition of autophagy can lead to increased autophagosome accumulation, depending on where in the pathway the perturbation occurs.
The Autophagy LC3 HiBiT Reporter Assay System can be used in three detection modules to unambiguously quantitate autophagic flux. First, the reporter assay measures autophagic flux by monitoring total levels of the LC3-based reporter in a plate-reader based assay. Cells stably expressing the autophagy reporter and treated with stimulators of autophagy will show a decreased luminescent signal. Treatment with inhibitors of autophagy results in a buildup in the level of LC3-based reporter and thus a higher luminescent signal. Second, the HiBiT tag on the LC3 reporter can be used with the Nano-Glo® HiBiT Blotting System to monitor the molecular weight change from LC3-I to LC3-II. This system replaces cumbersome western blots with a faster and simpler protocol. Finally, the HaloTag® portion of the reporter can be used with fluorogenic dyes to image the location of LC3 within the cells. Using a single reporter to perform quantitative plate-reader based assays, along with complementing imaging and blotting assays, allows versatile analysis of the autophagic pathway.