Luciferase Reporter Assay for Deciphering GPCR Pathways
Zhijie Cheng, Aileen Paguio, Denise Garvin, Pete Stecha, Keith Wood and Frank Fan
Promega Corporation
Abstract
G protein-coupled receptors (GPCRs), also known as 7TM receptors, constitute the largest superfamily of cell surface receptors. These receptors play key roles in regulation of a wide variety of physiological processes, including cognition, metabolism, inflammation, immunity and cell proliferation, as well as development. As a result, GPCRs have been popular drug targets; 40% of approved drugs (or drugs currently on the market) directly target GPCRs. Despite the large number of GPCRs that are available as potential drug targets, currently approved drugs only address a small portion of the known receptors; hundreds of other compounds are under development. Thus, this target class remains an area of strong interest in drug discovery.
Introduction
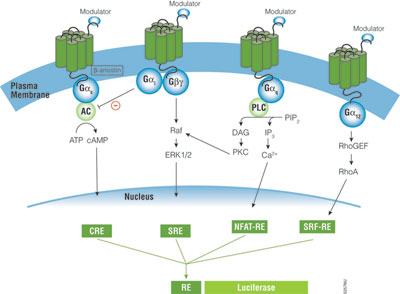
A primary function of GPCRs is transmission of extracellular signals across the plasma membrane via intracellular coupling with heterotrimeric G proteins. In addition, emerging evidence indicates GPCRs can signal independently of G proteins. Heterotrimeric G proteins are classified into four subfamilies based on their Gα subunit, Gs, Gi, Gq and G12. Upon activation of GPCRs, the Gα protein subunit dissociates from the βγ-dimeric subunit, initiating a cascade of downstream second messenger pathways that eventually induces gene transcription by various response elements (RE), including cAMP response element (CRE), nuclear factor of activated T-cells response element (NFAT-RE), serum response element (SRE) and serum response factor response element (SRF-RE) (Figure 1).
Despite the large number of GPCRs that are available as potential drug targets, currently approved drugs only address a small portion of the known receptors; hundreds of other compounds are under development.
The G protein coupling profile has not been fully characterized for many GPCRs. Some GPCRs only couple to one type of G protein (e.g., Gs or Gi), but many GPCRs couple to a broad range of G-protein families, such Gi/G12, Gq/G12, or Gi/Gq/G12,(1) . Current methods employed in GPCR screening programs measure G protein signaling by determining change in second messengers such as cAMP, inositol triphosphate (IP3) and intracellular Ca2+ mobilizations, which often demands setting up different assay platforms and requires specialized instrumentation for each pathway and can be costly. Reporter gene assays offer a simple solution for the study and characterization of receptor/G protein coupling, because reporter vectors with specific response elements (CRE, NFAT-RE, SRE, SRF-RE) are available for Gs, Gi, Gq and G12 pathways (Figure 1).
Rapid Response™ Reporters Improve Assay Dynamics
Reporter gene systems are used widely for studying gene regulation in response to GPCRs. There are many reporter genes available, including alkaline phosphatase, β-galactosidase, β-lactamase, green fluorescent protein and luciferase. Among them, luciferase is the reporter of choice especially in high-throughput screening due to its sensitivity, broad dynamic range, lack of endogenous activity and inherently low interference from compound libraries (2). The pGL4 Vectors containing minimal promoter and Rapid Response™ luciferase have been shown to improve the responsiveness of cAMP response element to Gs-coupled receptor. We further constructed SRE and SRF-RE reporter in various pGL4 Vectors and transiently expressed these reporters in HEK293 cells. By adding protein degradation sequences hPEST and hCL to the C-terminus of luciferase, both SRE- and SRF-RE reporters gave a larger induction response (75- to 120-fold) with a lower basal activity than did traditional luciferase without degradation signals (Figure 2). In addition, the destabilized luciferase response was rapid, reaching a maximum induction in 4–6 hours.
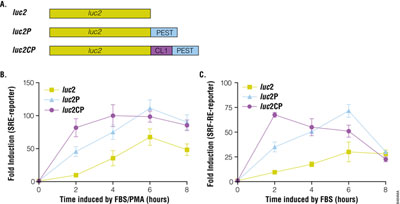
The pGL4.33[luc2P/SRE/Hygro] Vector and pGL4.34[luc2P/SRF-RE/Hygro] Vector contain SRE and SRF-RE, respectively, upstream of the minimal promoter and Rapid Response™ luciferase. SRE is known to respond to ternary complex factor (TCF)-dependent MAP kinase pathway, while SRF-RE, a mutant form of SRE lacking TCF binding domain, is designed to respond to SRF-dependent and TCF-independent pathways, such as RhoA activation. GPCRs, particularly those coupled with Gi and Gq can activate the MAPK pathway and induce transcriptional activation of SRE, while GPCRs coupled with G12 family are known to activate Rho guanine nucleotide exchange factors (RhoGEFs), which leads to activation of RhoA and transcriptional activation of SRF-RE (3).
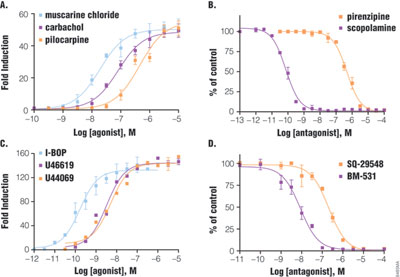
HEK293 cells transiently transfected with pGL4.33 and pGL4.34 reporter vectors show a large dynamic range of luciferase expression upon activation of GPCR (Figure 3). Cells expressing m3 muscarinic receptor (Gq-coupled) and SRE reporter showed 50-fold induction after six hours of treatment with various agonists. Among the agonists, muscarinic chloride was the most potent, with an EC50 of 19nM, followed by carbachol, then pilocarpine. Two antagonists were tested in the presence of carbachol: scopolamine with an IC50 of 0.07nM was much more potent than pirenzipine. Similarly, double transfection of SRF-RE with thromboxane A2 receptor (G12-coupled) showed a dose-response curve with agonists (120-fold induction) and antagonists. The potency ranking for agonist is I-BOP (EC50 =0.16nM) > U46619 = U44069 and for antagonist, as BM-531 > SQ-29548. The orders of potency ranking are consistent with previous reports, indicating that results from luciferase reporter assay reflect biologically relevant conditions (4–5).
High-Quality Assay for GPCR High-Throughput Screening
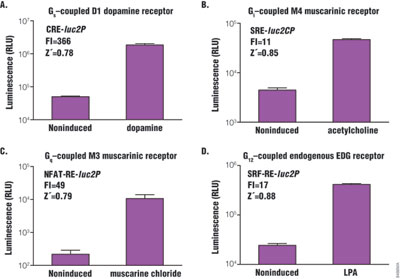
Z´-factor is the major statistical parameter used to evaluate assay performance in high-throughput screening for drug discovery. The Z´-factor is determined as follows: Z´ = 1 – [(3 × SD high + 3 × SD low) / (average high – average low)]. The closer the Z´-factor is to 1, the better the assay quality. In general, values >0.5 are acceptable for HTS. Four GPCRs were chosen: D1 dopamine receptor, m4 muscarinic receptor, m3 muscarinic receptor and endogenous EDG receptor. All are known to couple with Gs, Gi, Gq and G12 subfamily, respectively, and co-express stably (CRE and NFAT-RE) or transiently (SRE and SRF-RE) with the corresponding reporter vector in HEK293 cells. Cells were then stimulated by agonist, and Z´-factor values were determined to evaluate assay performance (Figure 4). All assays showed Z´-factor values significantly higher than 0.5 in a 384-well plate format, and the response dynamic (fold induction) ranged from 11- to 366-fold.
Using Luciferase Reporter Assay to Decipher GPCR Downstream Signaling Pathways
GPCRs can couple to multiple G proteins in one cell type, or couple to different G proteins in cell type-dependent manner. Also, more and more evidence indicates that the same receptor can activate multiple signaling pathways with different potencies depending on the ligand, which binds to the receptor orthosterically or allosterically (6). This has great potential for developing pathway-specific drugs that increase efficacy and reduce side effects, and add another dimension in assay complexity to compound screening. Despite significant interest, progress has been slow to include receptor/G-protein profiling in current drug screening practice, partly due to the significant burden of setting up multiple assay platforms.
Despite significant interest in receptor/G-protein profiling, progress has been slow to include GPCRs in current drug screening practice.
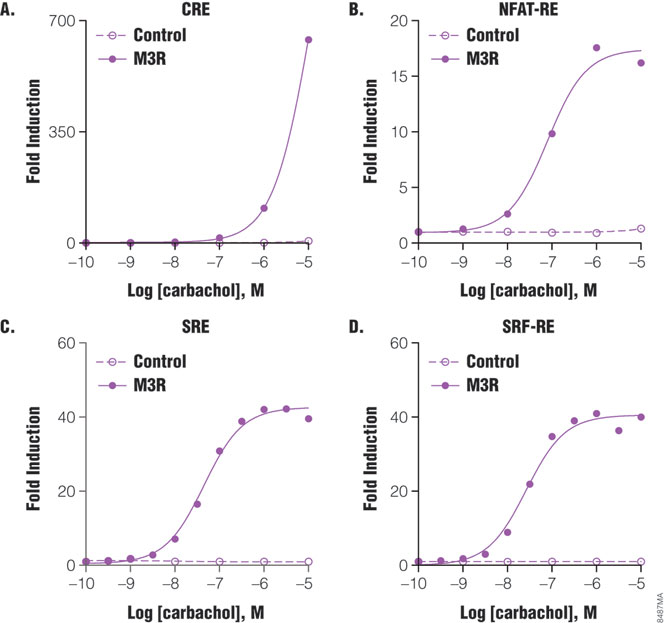
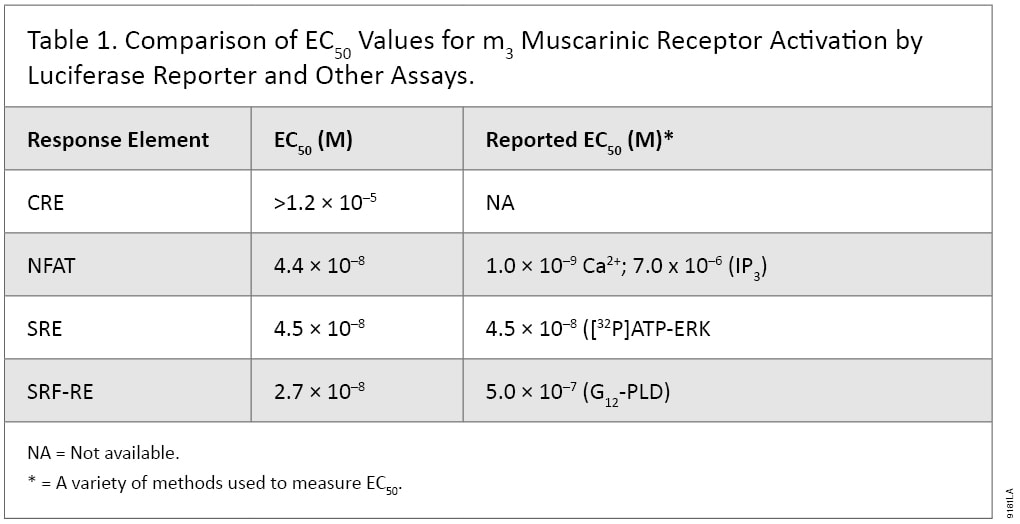
The Luciferase Reporter Assay is simple, sensitive and easy to set up. More importantly, it can measure Gs, Gi, Gq and G12 pathways in a single assay format, and therefore is well suited to pharmacological studies. For example, m3 muscarinic receptor is known to couple with Gq and G12 pathways in multiple cell types. When co-expressing with individual reporter vectors, m3 muscarinic receptor activation by agonist carbachol-induced dose-dependent expression of luciferase by response elements CRE, NFAT-RE, SRE and SRF-RE with different potencies (Figure 5). As indicated in Table 1, NFAT-RE- and SRE- reporter vector assays showed similar EC50 values for receptor activation, confirming that activation of m3 muscarinic receptors leads to increased intracellular calcium concentration and MAP kinase activation; both are known to be mediated by Gq protein. The EC50 value of the SRF-RE reporter assay is within the same log range as the values of the SRE and NFAT-RE reporter assays, indicating that m3 muscarinic receptors couple with G12 protein as efficiently as with Gq. In contrast, EC50 value of the CRE reporter assay is almost three logs higher than those by other reporter assays tested, in agreement with previous reports that m3 muscarinic receptors couple with Gs in a less efficient manner (7). All the EC50 values for different signal pathways via m3 muscarinic receptors reporter assays showed similar or better sensitivities to those achieved using other detection technologies (Table 1). Thus, luciferase reporter technology is able to study G-protein activation profiles in a single assay format.
Summary
Here we have shown that Rapid Response™ luciferase reporter is well suited for studying GPCR signaling pathways and for high-throughput screening in drug discovery. GPCR modulators can be readily evaluated for their potency as agonists or antagonists, and the results reflect biologically relevant conditions. Luciferase reporter assays are easily applied to high-throughput screening with high Z´-factor values. In a single-assay format, individual GPCR pathways (Gs, Gi, Gq and G12) can be measured using CRE-, NFAT, SRE- and SRF-RE reporter vector assays, respectively.
References
- Wettschureck, N. and Offermanns, S. (2005) Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–204.
- Fan, F. and Wood, K. (2007) Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 5, 127–36.
- Hill, C.S. and Treisman, R. (1995) Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 14,5037–47.
- Offermanns, S. et al. (1994) Transfected muscarinic acetylcholine receptors selectively couple to Gi-type G proteins and Gq/11. Mol. Pharmacol. 45, 890–8.
- Zhang, L. et al. (2006) The G12 family of G proteins as a reporter of thromboxane A2 receptor activity. Mol. Pharmacol. 69, 1433–40.
- Christopoulos, A. and Kenakin, T. (2002) G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54, 323–74.
- Nahorski, S.R., Tobin, A.B. and Willars, G.B. (1997) Muscarinic M3 receptor coupling and regulation. Life Sci. 60, 1039–45.