Making Sense of Genetic Reporter Data by Assay Multiplexing
Mark Bratz, Brad Hook and Trista Schagat
Promega Corporation
Publication Date: November 2012
Abstract
Genetic reporter assays allow researchers to investigate the activation and repression of transcription dictated by various pathways in cellular systems. Compounds that activate and repress cellular pathways are very important in modern scientific research. Reporter assays are implemented by introducing a DNA construct with a pathway-specific sequence regulating expression of a reporter gene that is easy to assay. Often, this is a luciferase gene that will be expressed if the pathway is active. If the pathway is always active unless an inhibitor is introduced, the luciferase will be constitutively expressed as well. Changes in these reporter levels can then be correlated to changes in pathway activation. Compounds that are cytotoxic present a challenge to reporter assays because a reduction in luminescent signal may be a simple reduction in the number of viable cells. Therefore, various normalizing methods exist, such as the Dual-Glo® Luciferase Assay System and ONE-Glo™ + Tox Luciferase Reporter and Cell Viability Assay, to protect against misinterpreting data. In this report, we describe how the CellTox™ Green Cytotoxicity Assay was tested for its potential as a candidate for multiplexing with reporter assays. The IC50 values obtained from multiplexing these assays were not significantly different from IC50 values obtained using the reporter assay formats alone, and false reporter signal reductions, revealed by inversely concordant changes in viability.
Introduction
The CellTox™ Green Cytotoxicity Assay monitors cell health by detecting changes in membrane integrity. The assay employs DNA binding dye that is excluded from cells with intact membranes (Figure 1). If a membrane has been compromised, the dye can bind to the DNA of dead cells. DNA binding yields a marked increase in the fluorescent properties of the dye, and fluorescence is proportional to cytotoxicity. The dye is well tolerated by growing cultures and can be diluted directly into culture medium and added directly to the cell culture at plating or compound dosing. This “no-step” approach allows researchers to monitor cell health throughout the experiment. Alternatively, CellTox™ Green Dye can be added in an endpoint format after compound exposure. Because this assay is based on differential membrane permeability and DNA staining, it does not depend on an activity-based biomarker. Activity-based markers tend to degrade after a cytotoxic event leading to a loss of signal if the cytotoxicity assay is not administered when maximal amounts of biomarker are present. DNA is a more stable biomarker that yields a long-lasting signal.
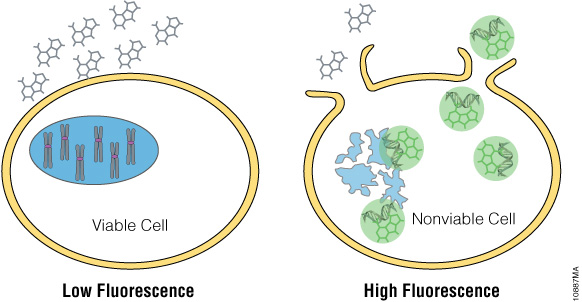
Figure 1. The CellTox™ Green Cytotoxicity Assay. CellTox™ Green Dye is excluded from viable cells, but binds to DNA from dead cells with compromised membrane integrity.
Multiplexing is a convenient and efficient way to maximize the amount of data collected and can improve data content while minimizing the number of experiments needed (1). In particular, multiplexing cytotoxicity assays with a reporter assay is extremely advantageous. Reporter assays allow researchers to investigate a wide variety of areas in gene regulation (2). Many different tools exist for studying multiple different regulation pathways. Multiplexing these gene regulation experiments with a cytotoxicity assay allows researchers to verify if changes in luminescent signal result from changes in the genetic pathway or a cytotoxic event. Multiplexing has been shown to eliminate faulty interpretation of experimental data. In this report, we show that the CellTox™ Green Cytotoxicity Assay can be multiplexed with many luciferase reporter assays to improve data interpretation.
Methods
Materials Required
- CellTox™ Green Cytotoxicity Assay, Cat.# G8741
- Multimode plate reader, e.g., GloMax®-Detection System, Cat.# GM3000
- CellTox™ Green Cytotoxicity Assay, Cat.# G8741
- ONE-Glo™ Luciferase Assay System, Cat.# E6110
- Bright-Glo™ Luciferase Assay System, Cat.# E2610
- Steady-Glo® Luciferase Assay System, Cat.# E2510
- Nano-Glo® Luciferase Assay System, Cat.#N1110
- Renilla-Glo™ Luciferase Assay System, Cat.# E2710
- Nuclease-Free Water, Cat.# P1193
- stably transfected HEK293 cells with constitutive CMV-Firefly Luciferase production
- stably transfected HEK293 cells with constitutive CMV-Renilla Luciferase production
- stably transfected HEK293 cells with constitutive CMV-NanoLuc™ Luciferase production
- stably transfected HEK293 cells with constitutive CMV-NanoLuc™-PEST Luciferase production
- terfenadine, Sigma Cat.# T9652-5G
- ionomycin calcium salt from Streptomyces conglobatus, Sigma Cat.# I0634-1MG
Evaluating the CellTox™ Green Cytotoxicity Assay in Multiplex with a Luciferase Reporter Assay
HEK-293 cells constitutively expressing various luciferase reporters were plated at 5,000 cells/well in 50μl of DMEM in the inner wells of the 96-well plates to eliminate edge effects. A total of 50μl of compound diluted in DMEM was added to the cells. The cells and compound were incubated at 37°C + 5% CO2 for 5–6 hours. A plate without CellTox™ Green was assayed as a control to determine the potential deleterious effects of adding CellTox™ Green to the assay. CellTox™ Green was tested with two protocols: The “no-step” protocol with 4μl added to every 500μl of compound dilution as well as the endpoint protocol where 20μl of CellTox™ Green was diluted with 2ml of Assay Buffer and 20μl of the resulting dilution was added to the cells. Endpoint plates were incubated for 15 minutes at room temperature to allow for interaction with released DNA. Endpoint data were similar to the “no-step” protocol data and are not reported here. The cytotoxicity was determined by measuring the fluorescence before “-Glo” reagents were added to the reactions. The respective “-Glo” reagent was then added to the wells and incubated at room temperature for the suggested time in the technical manuals. After incubation, luminescence was measured.
Results
To investigate the applicability of multiplexing CellTox™ Green with the portfolio of reporter assays that Promega offers, the compound terfenadine (200μM to 0.78μM) was chosen due to its known cytotoxic effects. Table 1 and Figure 2 show the results of all the reporter assays multiplexed with the CellTox™ Green Assay. There were no significant differences between the IC50 values determined with and without CellTox™ Green Dye present. There was a slight decrease in the overall luminescent signal due to the introduction of CellTox™ Green. A point of interest in these data was the NanoLuc® luciferase response to terfenadine. The NanoLuc® luminescent signal does not decrease as much the signal generated from the other luciferase reporters. The NanoLuc-PEST™ luciferase has a much more pronounced decrease in luminescent signal as the cells are killed. We believe that reduction in the fold-change in signal is due to the stability of the NanoLuc® luciferase. This evolved luciferase protein is more stable outside of the cell (3) . Therefore, once the cells are killed, the NanoLuc® luciferase will retain activity in the cell medium, reducing observed luciferase signal loss. This phenomenon illustrates the importance of carefully selecting the appropriate luciferase reporter for your experimental system.
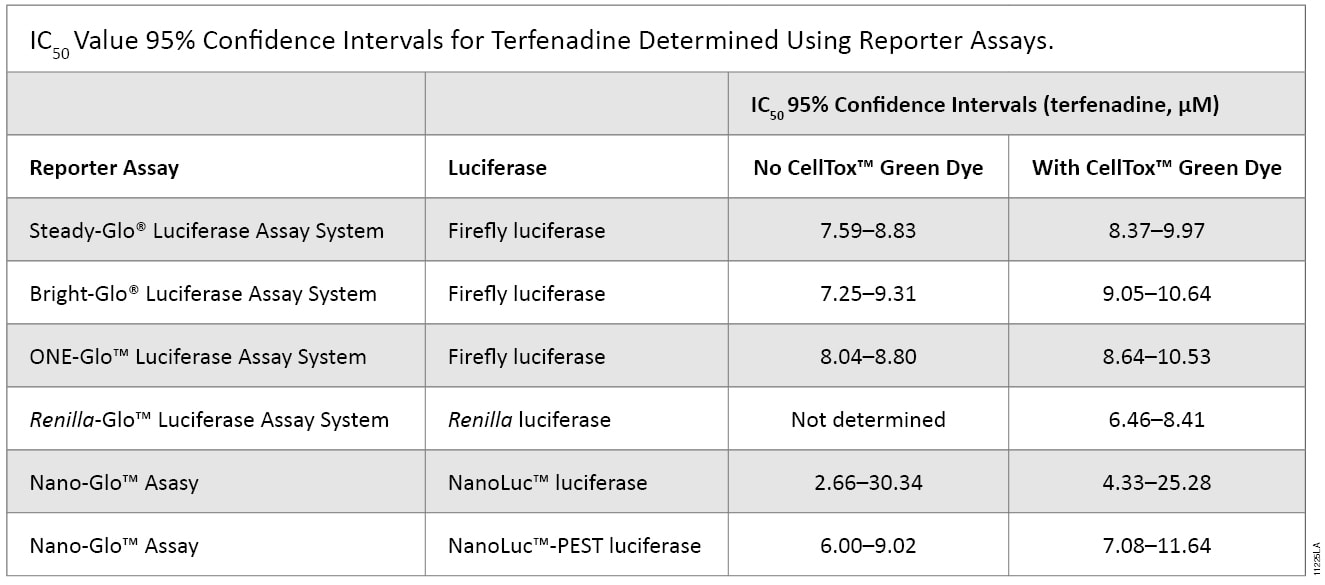
Table 1. The IC50 values of the reporter assays are not significantly different in the presence or absence of CellTox™ Green Dye. The table shows the statistical 95% confidence intervals obtained for the cytotoxic agent terfenadine, calculated using GraphPad Prism® 5 software. The calculations are based on replicates of six.The IC50 values of the reporter assays are not significantly different in the presence or absence of CellTox™ Green Dye. The table shows the statistical 95% confidence intervals obtained for the cytotoxic agent terfenadine, calculated using GraphPad Prism® 5 software. The calculations are based on replicates of six.
The cells expressing Renilla luciferase showed a pronounced decrease in the luminescence curve and a correspondingly sharp increase in the fluorescence curve. To test if this was just a compound effect on this particular cell line, ionomycin was used to treat the cells instead of terfenadine. The ionomycin was diluted threefold from 100μM to 0.02μM. Figure 3 illustrates that the abnormal curves were not seen with the ionomycin experiment, and yet the IC50 values were still not significantly different. The multiplexed data show that cytotoxic events can reduce the reporter signal, and therefore, a cytotoxicity assay such as CellTox™ Green becomes essential for correct data interpretation.
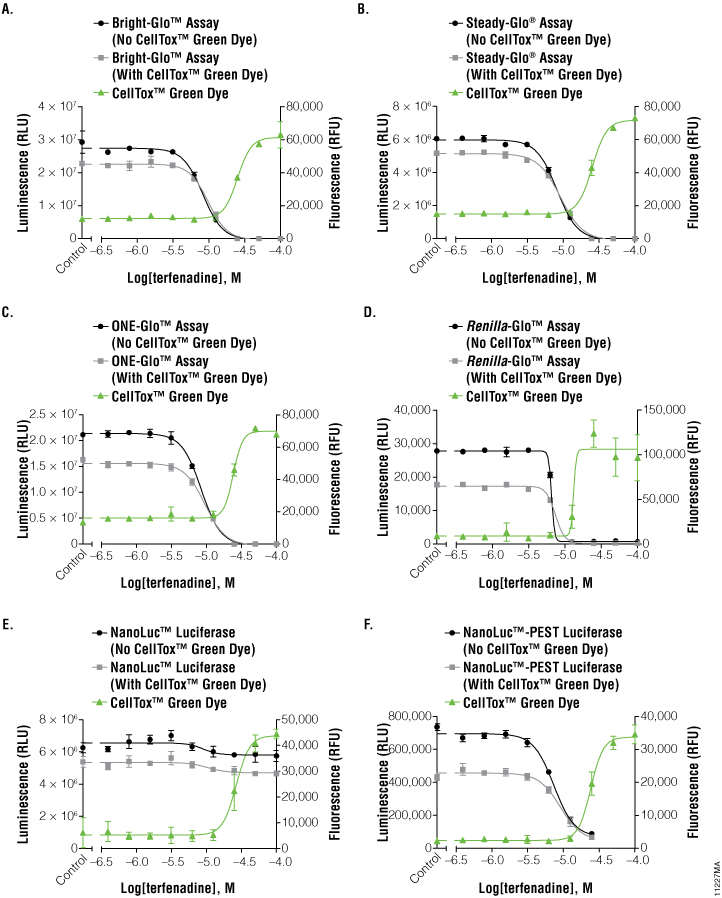
Figure 2. Graphs of fluorescent and luminescent data from the reporter assays completed with the compound terfenadine.
The graphs reveal the inverse concordance that is expected from the multiplex of the CellTox™ Green Cytotoxicity Assay (fluorescence, RFU) and the luciferase reporter assay (luminescence, RLU). Luminescent signal declines as cells die and produce less protein. Fluorescent signal rises as the cell membrane becomes compromised, allowing the CellTox™ Green Dye to bind to the DNA. Panels A, B, and C are the Bright-Glo™, Steady-Glo®, and ONE-Glo™ Assays performed on HEK293 cells producing firefly luciferase constitutively; panel D is the Renilla-Glo™ assay performed on HEK293 cells producing Renilla luciferase constitutively; panels E and F are the Nano-Glo™ assays performed on HEK293 cells producing either NanoLuc™ luciferase or NanoLuc™-PEST luciferase constitutively, as indicated. Data are the average and standard deviation of six replicates.
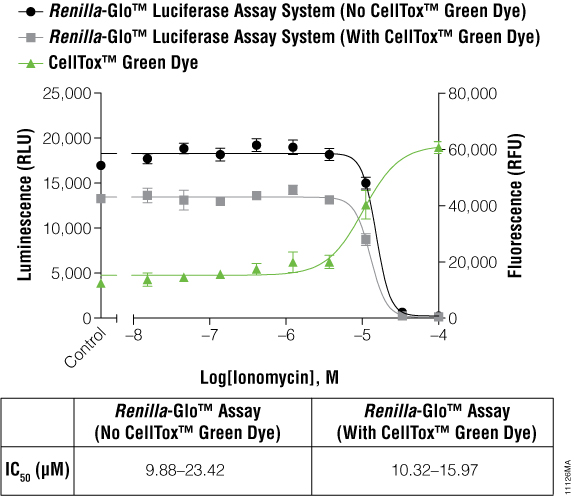
Figure 3. IC50 values obtained for ionomycin.
The inverse concordance is shown again between the luminescent and fluorescent data. The luminescence is reduced due to the multiplex of the assays, but IC50 values are not significantly different. IC50 values are reported as 95% confidence intervals as calculated using GraphPad Prism® 5 software on replicates of six.
Summary
In summary the data presented here show that the CellTox™ Green Cytotoxicity Assay is compatible as a multiplex assay with luciferase reporter assays. Although a modest reduction in luminescence values is observed when multiplexing with CellTox™ Green, most likely from modest absorption of the luminescent signal by the fluorescent dye, this signal reduction does not affect assay-generated IC50 values. The luminescent signal was, on average, 15.9% lower than if CellTox™ Green Dye had not been added. The ability to add the CellTox™ Green Dye at dosing or plating (“no-step” protocol) allows researchers to monitor cytotoxicity throughout an experiment, providing valuable data about cell health and allowing more successful interpretation of reporter assay data.
References
- Hooper, K. (2011) Multiplexing cell-based assays: Get more biologically relevant data. Promega PubHub.
- Allard, S.T.M. and Kopish, K. (2008) Luciferase reporter assays: Powerful, adaptable tools for cell biology research. Cell Notes 21, 23–6.
- Hall, M.P. et al. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848–57.
Related Products
CellTox™ Green Cytotoxicity Assay
Visit the product page to learn more about the CellTox™ Green Assay, view more data or download the protocol.