Detection of HIF1α Expression at Physiological Levels Using NanoLuc® Luciferase and the GloMax® Discover System
Promega Corporation
Publication Date: Nov. 2014 (tpub_156)
Abstract
Here we demonstrate the utility of NanoLuc® Luciferase for measuring gene expression at physiologically relevant levels using hypoxia inducible factor 1 alpha expression (HIF1α) as an experimental system and demonstrate detection with the GloMax® Discover System.
The Need for a More Sensitive Reporter System
Reporter genes have long been used as a method to assay for expression of a particular gene of interest. Constructs that result in overexpression of the gene may result in changes in the cellular response or other deleterious effects that can affect the interpretation of the results of the reporter assay. Thus, measuring gene expression at physiologically relevant levels becomes important. However, weakly expressing genes can be difficult to measure and can require a more sensitive system for analysis. With the availability of zinc-finger, TALEN and CRISPR/Cas genome editing methods and small, sensitive reporters like NanoLuc® Luciferase, designing reporter assays to faithfully detect endogenous gene expression is increasingly achievable. In this report, we examine the ability of one such reporter assay to detect HIF1α (hypoxia inducible factor 1 alpha subunit) expression at physiological levels using NanoLuc® Luciferase and the GloMax® Discover System.
Induction of a Cellular Response to Hypoxic Stress
HCT-116 cells containing an in-frame fusion of HIF1α to the NanoLuc® reporter gene [X-MAN™ HIF1A (+NanoLuc®/+) protein reporter] (1) were grown in complete medium (RPMI 1640 media + 10% FBS + 300µg/ml G418). Cells were washed with PBS, trypsinized, and diluted to 2.22 × 105 cells/ml in complete medium (20,000 cells/90µl). Cells were then serially diluted twofold in complete medium to a final concentration of 10K cells/90µl, 5K cells/90µl, and 2.5 × 103 cells/90µl. A total volume of 90μl of cells was plated per well into 2 solid-white 96-well plates for each concentration. The cells were incubated for ~20 hours in a 37°C, 5% CO2 incubator.
A 10mM stock solution of 1, 10-phenanthroline was serially diluted in DMSO, and a 250mM stock solution of CoCl2 was serially diluted in Nuclease-Free Water, including vehicle-only controls for each compound. The dilutions were further diluted 1:10 in RPMI 1640 media + 10% FBS to a 10X concentration. A total volume of 10µl of each dilution was added to each cell number in quadruplicate 96-well plates and incubated for 3 hours in a 37°C, 5% CO2 incubator. Plates were prepared in duplicate. After a 3-hour exposure time, 100μl of Nano-Glo® Reagent was added to each well. The plates were mixed briefly, incubated at room temperature for 10 minutes, and read on the GloMax® Discover System.
HIF1α Expression Detected at Physiological Levels
Upon treatment with phenanthroline and CoCl2, HCT-116 cells containing the in-frame fusion of HIF1α to the NanoLuc® reporter gene can be monitored for inducible changes in HIF1α. The cells responded to both compounds in a dose-dependent manner as evidenced by the increase in NanoLuc® Luciferase expression as the concentrations of the compounds were increased (Figures 1 and 2). For phenanthroline, induction increased up to ~12.5µM. Higher concentrations resulted in no increase in luciferase induction. For CoCl2, induction increased up to ~92.6µM. Higher concentrations resulted in a pronounced decrease in luciferase induction likely as a result of compound toxicity and cell death. Expression of uninduced NanoLuc® Luciferase at the various cell numbers are also shown in Figure 3, indicating the ability of the GloMax® Discover System to detect HIF1α- NanoLuc® Luciferase at physiological levels. During the validation of this cell line, cells containing a construct in which HIFα was replaced (not fused) with NanoLuc® Luciferase were tested to so that the protein is being properly regulated and that induction is not a result of transcriptional changes (1) .
 Figure 1. Induction of NanoLuc® Luciferase in HCT-116 cells containing the in-frame fusion of HIFα to the NanoLuc® reporter gene by CoCl2.
Figure 1. Induction of NanoLuc® Luciferase in HCT-116 cells containing the in-frame fusion of HIFα to the NanoLuc® reporter gene by CoCl2. Various cell numbers were treated by the compound for 3 hours. Luminescence was quantified on a GloMax® Discover System. RLUs plotted were background subtracted based on the vehicle and no-cell controls.
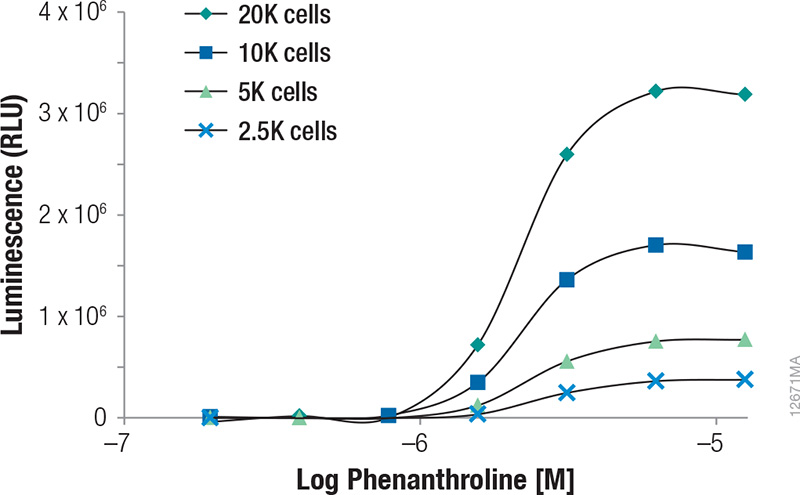 Figure 2. Induction of NanoLuc® Luciferase in HCT-116 cells containing the in-frame fusion of HIFα to the NanoLuc® reporter gene by 1,10 Phenanthroline.
Figure 2. Induction of NanoLuc® Luciferase in HCT-116 cells containing the in-frame fusion of HIFα to the NanoLuc® reporter gene by 1,10 Phenanthroline. Various cell numbers were treated by the compound for 3 hours. Luminescence was quantified on a GloMax® Discover System. RLUs plotted were background subtracted based on the vehicle and no-cell controls.
 Figure 3. Physiological levels of NanoLuc® luciferase in HCT-116 cells containing the in-frame fusion of HIF1α to the NanoLuc® reporter gene at various cell numbers.
Figure 3. Physiological levels of NanoLuc® luciferase in HCT-116 cells containing the in-frame fusion of HIF1α to the NanoLuc® reporter gene at various cell numbers. Luminescence was quantified on a GloMax® Discover System. RLUs plotted were background subtracted based on the no-cell controls.
Summary
The GloMax® Discover System coupled with the NanoLuc® luciferase technology provide superior luminescence sensitivity and dynamic range for both strongly and weakly expressing bioluminescent reporters. Cells containing the in-frame fusion of HIF1α to the NanoLuc® reporter gene can be monitored for inducible changes in HIF1α using the GloMax® Discover, while the sensitivity of the instrument allows for the ability to detect expression of uninduced HIF1α at physiological levels with as few as 2,500 cultured cells. The design of reporter assays to detect endogenous levels of NanoLuc® Luciferase-tagged proteins are becoming increasingly achievable with the advent of genome editing tools like zinc-finger, TALEN and CRISP/Cas9 gene editing methods.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Wieczorek, D. Detection of HIF1α Expression at Physiological Levels Using NanoLuc® Luciferase and the GloMax® Discover System. [Internet] Nov. 2014 (tpub_156). [cited: year, month, date]. Available from: https://worldwide.promega.com/resources/pubhub/tpub_156-detection-of-hif1alpha-expression-at-physiological-levels-using-nanoluc-luciferase/
American Medical Association, Manual of Style, 10th edition, 2007
Wieczorek, D. Detection of HIF1α Expression at Physiological Levels Using NanoLuc® Luciferase and the GloMax® Discover System. Promega Corporation Web site. https://worldwide.promega.com/resources/pubhub/tpub_156-detection-of-hif1alpha-expression-at-physiological-levels-using-nanoluc-luciferase/ Updated Nov. 2014 (tpub_156). Accessed Month Day, Year.
GloMax, NanoGlo and NanoLuc are registered trademarks of Promega Corporation.
 Learn what new research the unusually bright NanoLuc® Luciferase can enable.
Learn what new research the unusually bright NanoLuc® Luciferase can enable.