SARS-CoV-2 and the Antibody Response in COVID-19 Patients
By: Emily A. Teslow, Ph.D., Medical Affairs • August 2020
Immunology Overview
The Humoral Immune Response
The human immune system consists of various cellular differentiation pathways, which play distinct functions in responding to the presence of foreign antigens and micro-organisms in the body. Initial responses to foreign pathogens, such as viruses or bacteria, typically result in an early innate immune response1. A portion of this response is mediated by cells bearing specific pattern recognition receptors, which detect foreign antigens on the surface of a pathogen, released by the pathogen, or released through the interaction of the pathogen and host cells1. If the innate response fails to resolve an infection, an adaptive immune response may be initiated, where white blood cells or lymphocytes, with receptors capable of binding and detecting a specific foreign antigen, will go through clonal amplification and further maturation processes1. An important mediator of this response occurs via lymphocytic cells called B-cells, which characteristically mature in the bone marrow1. During a primary innate immune response, naïve B-cells will recognize foreign antigens in the body. Along with additional stimuli, this activates the B-cells in order to further stimulate their differentiation into effector cells types such as plasma cells (Figure 1). Plasma cells secrete Y-shaped proteins, called antibodies, into the extracellular environment1. B-cells may also further differentiate into memory B-cells, which may survive for decades in order to repeatedly produce an antibody-mediated response upon reinfection with a specific antigen1. This process of B-cell maturation, production of plasma and memory B-cells, and secretion of specific antibodies is part of the humoral immune response1.
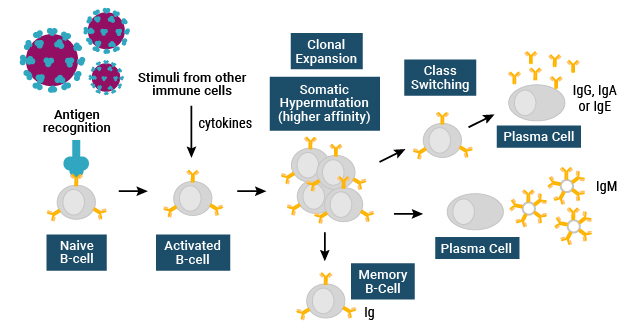
Figure 1. The humoral immune response.
During the humoral immune response, the secretion of antibodies by plasma cells into the extracellular environment may contribute to immunity in three ways. The first is through neutralization, through which an antibody will bind to an antigen on a foreign pathogen and block it from invading or attaching to a host cell, thus impeding further infection1. Another method is through a process called opsonization, in which antibodies attach to a foreign pathogen and induce phagocytosis or engulfment by innate immune cells. This leads to degradation of the pathogen, thus removing it from the body1. Lastly, antibodies may attach to a pathogen and activate the complement system, a very early innate immune pathway that assists in degradation of the pathogen1.
Antibody Diversity
All antibody-mediated immune responses are dependent on the ability of an antibody to bind specific antigens to recognize a foreign pathogen. However, not all antibodies are the same. Antibodies, also called immunoglobulins (Ig), are categorized into 5 different Ig classes: IgM, IgD, IgG, IgA, and IgE (Figure 2)2.
Each type of immunoglobulin molecule is comprised of a combination of constant regions and antigen binding sites. Antigen binding sites are highly variable regions represented at the top of the two arms of the Y-shaped antibody molecule. The human genome codes for millions of antigen binding sites. The constant regions, which make up the rest of the molecule, give rise to different antibody classes and provide different effector functions. They attract and stimulate different types of immune cells and some of them can activate the complement system1.
IgG and IgM are the most abundant classes of antibodies found in human serum, accounting for 75-85% and 5-10% of all Ig in serum respectively3. Secreted IgM antibodies are large pentamer molecules with 10 antigen binding sites, which typically show lower affinity for the target antigens1. IgM antibodies are produced early in the adaptive immune response and are great at activation of innate immune responses3. IgG, IgE, and IgD antibodies are monomers with two antigen binding sites. IgA antibodies may be found as monomers in blood or as dimers in mucosal interstitial spaces.
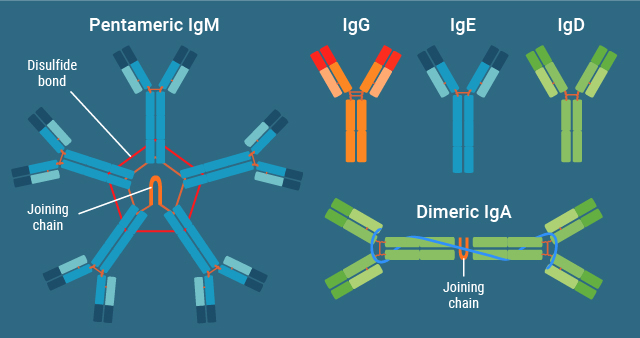
Figure 2. Immunoglobulin subclasses and their structures.
Before a naïve B-cell experiences an antigen, it will express cell surface receptor forms of IgM and IgD. Each B-cell produces an IgM-like receptor with only a single type of antigen binding site. If the B-cell receptor binds to an antigen, it triggers proliferation of that specific type of B-cell and its differentiation into plasma cells, which secrete the IgM antibody. During B-cell proliferation, enzymatic processes lead to somatic mutation of specific regions of the gene coding for the antibody. Some of these mutations will increase the affinity of the B-cell receptor for the antigen. Cells expressing a receptor with increased affinity will be stimulated to proliferate, while those with low-affinity receptors will die through apoptosis. As a result, different populations of B-cells expressing receptors with different affinities for the antigen will be produced. This process is referred to as clonal expansion.
During clonal expansion, a process referred to as class switching may also occur to produce different Ig subclasses, after which further mutations will continue to increase the affinity of the antibody for its antigen1. Thus, IgG will typically show higher affinity for an antigen than its related IgM, even though they may share the same specificity for that antigen1.
Seroconversion and Serologic Data
The immune system produces and circulates antibodies, specifically IgM, following an acute phase of disease. This is referred to as seroconversion, and it indicates that a recovery or convalescent phase of disease has begun. Once seroconversion occurs, antibodies may be detectable in the blood by antibody tests, or serological assays. Serologic data derived from antibody subclass and titer provides a history of infection and can be used to determine the nature of an infection, monitor population-based prevalence of disease, assess disease stage, guide vaccine development and identify an infectious agent such as a virus1,3. In addition, serologic data can be used to better understand the time course of an antibody mediated response to expand biological knowledge. This may be assessed through measuring titers of specific antibodies for different antigens and Ig classes over time using serological assays3. For example, in a typical immune response, IgM is generally detected during the first 3-14 days following initial infection3. IgG, IgA or IgE production may begin later, following a class switching event3. IgG and IgM concentrations will eventually peak and then either slowly decrease or plateau, in a time frame that could be specific to the pathogen and disease burden3. These assays can also be combined with other tests, which demonstrate the ability of the antibodies to neutralize virus3. Response to infectious disease agents may vary, so understanding the immune response for new and emerging pathogens is essential for determining the value and application of a serological test.
COVID-19 and the Immune Response to SARS-CoV-2
Serological Assays for SARS-CoV-2
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), formerly referred to as 2019-nCoV4, is the novel infectious disease agent which causes COVID-19. SARS-CoV-2 serological assays used to detect antibodies in the blood (ex: serum or plasma)5, can be useful for identifying exposed persons who have mounted an immune response but may be asymptomatic, show negative results in PCR testing, produce a low viral load, or present later in the course of disease. They may also be useful when respiratory sampling is not possible6. However, SARS-CoV-2 serological assays are not currently recommended for diagnosis or measurement of active infection in the United States (U.S.)6. According to the U.S. Food and Drug Administration (FDA), serological tests should only be ordered by physicians who are familiar with their use and limitations6.
The application of serological testing for COVID-19 disease is actively being studied. Other areas of potential use for SARS-CoV-2 serologic (antibody) testing include generating serologic data in order for researchers to study time-course of disease, supporting vaccine development strategies3, assessing the number of exposed individuals within a population7, assisting the medical community in understanding the adaptive immune response to SARS-CoV-2, or identifying eligible donors of convalescent plasma as a possible treatment strategy for critically ill COVID-19 patients6,8.
The prevalence of SARS-CoV-2 infection in the U.S. population is currently unknown9. Serological assays, which detect the presence of antibodies to different viral proteins, may help assess prevalence of COVID-19 disease and determine whether an individual has been previously infected, but may not indicate whether an individual has developed immunity9.
Presently, numerous serology tests have been granted Emergency Use Authorization (EUA) by the FDA for SARS-CoV-2 antibody detection, which differ based on their performance characteristics9, the technological approaches they employ to detect antibodies and the class of antibodies they are capable of detecting in blood for different SARS-CoV-2 antigens5,10. For example, some serological assays may only detect IgG, while others detect both IgG and IgM, or all antibody classes. They may also detect different SARS-CoV-2 viral antigens5,10. Some tests have demonstrated greater sensitivity with combined IgG and IgM measurement11-16, and recent meta-analysis demonstrates that using antigens to viral spike (S) versus nucleocapsid (N) proteins may also increase sensitivity of serological tests13. The complete utility of these tests has yet to be determined for COVID-19 disease, as the world continues to obtain new information regarding the immune response to SARS-CoV-2.
For additional information about SARS-CoV-2 immune response and studies, visit the Promega Connections blog: www.promegaconnections.com
SARS-CoV-2 Antibody-mediated Immune Response
What we know about the SARS-CoV-2 antibody-mediated immune response is derived from a limited number of recent studies which have collected serologic data from COVID-19 patients, using both experimental and commercially available serological assays. Most of those assays do not currently hold Emergency Use Authorization from the FDA in the United States. From these studies, it has been demonstrated that after 2 weeks post-disease onset, the average seroconversion rate is between 90-100% for IgM-IgG antibodies produced against the SARS-CoV-2 S and N proteins14-19. (For more information on the SARS-CoV-2 viral structure and proteins please visit SARS-CoV-2 PCR and Serological Testing.) Serological data may be generated using several different types of assay technologies and formats, including antibody-mediated capture assays such as enzyme-linked immunosorbent assays (ELISA)16, 20-23, as well as antigen-mediated capture approaches such as chemiluminescent immunoassays (CLIA)11, 19, 24-26, immunofluorescence assays18, lateral flow assays27, luciferase immunoprecipitation assays (LIPS)28 and bioluminescent immunoassays. Using these types of serological assays, evidence of a humoral immune response in COVID-19 patients has been studied, and the profile of anti-SARS-CoV-2 antibodies in serum have been preliminarily described. Shown in Figure 3 is a graphical representation of the estimated antibody response in COVID-19 patients. Though many studies provide insights through preliminary serologic data, the question remains whether an antibody-mediated response may assist in clearing viral infection, and how clearance via antibodies might occur through various virus neutralizing immune responses.
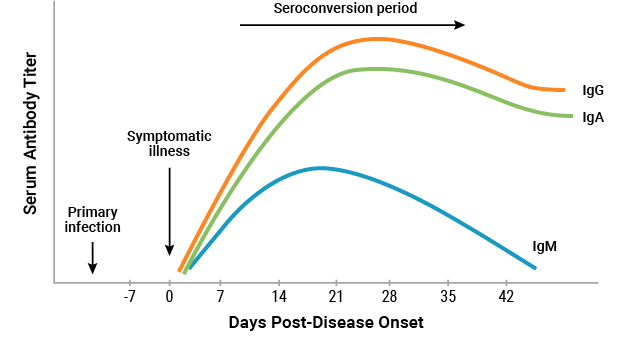
Figure 3. Graphical representation of the estimated antibody titers (IgG, IgA and IgM) produced by COVID-19 patients 6 weeks post-symptom onset (Illustration adapted from Lee et al37).
There remains a limited understanding of the magnitude and duration of antibody responses in COVID-19 patients with different disease burden and how factors such as comorbidities, age and genetic background impact immune response; however, there is mounting evidence that higher levels (titers) of total anti-SARS-CoV-2 antibodies may be associated with higher disease burden when detected 2 weeks post-symptom onset18,19,23-25. For example, Qu et al. demonstrated that critically ill patients in China produce higher titers of IgM and IgG antibodies to S and N proteins, and kinetically observed that IgM levels peaked later in disease course in COVID-19 patients who were critically ill versus patients with mild to moderate disease24.
Similarly, Zhao et al. observed higher IgM and IgG titers in critically ill COVID-19 patients23. However, this increase in serum antibodies did not coincide with a reduction in viral RNAs within nasopharyngeal samples of these critically ill patients23. Moreover, disease burden in this case was not associated with the levels of SARS-CoV-2 IgM antibodies produced to the S protein or IgG antibodies produced to the N protein23, suggesting that only certain antibodies may be associated with disease burden. In contrast, Long et al. found no association between IgG antibody production and clinical attributes of patients in China18, and Wӧlfel et al. demonstrate that in a small cohort of COVID-19 patients in Germany, the production of neutralizing antibodies in serum may not correlate well with clinical disease course25. This result is in conflict with that of Ni et al. who discovered that COVID-19 patients recently discharged from a hospital setting produced significant titers of virus neutralizing antibodies against the receptor binding domain (RBD) of the S protein, which was associated with production of virally targeted lymphocytes12. Likewise, an in vitro study demonstrated that antibodies against the SARS-CoV-2 S protein can inhibit entry into cells29, and a few other studies have observed that COVID-19 patients produce neutralizing antibodies to the RBD domain of the SARS-CoV-2 S protein30-35. Lastly, COVID-19 patients with severe acute infection have presented with higher plasma concentrations of pro-inflammatory cytokines, which contribute to disease-related mortality32. However, the ways that this attribute may impact the effectiveness of an antibody-response is currently unknown. Overall, there remains an unclear understanding of the relationship between the humoral immune response and COVID-19 disease, and as such it remains to be elucidated through further research which antigens and antibody classes may be associated with a more robust immune response, how that response occurs, and whether or not that response coincides with complete disease recovery in COVID-19 patients.
With the continuing development of technologies used for serological measurement of SARS-CoV-2 antibodies, the world may gain a better understanding of the emerging disease, COVID-19. This type of information is both useful to the medical community in the interim for monitoring patient response, and through research which has the potential to expand our knowledge of the body’s natural responses to the novel SARS-CoV-2 virus so that we may be able to discover new ways to control disease within the population.
Acknowledgments: Drafts of this article were edited by Kara Chamberlain, Ph.D. and Annette Burkhouse, Ph.D.
References:
1Murphy, K., Travers, P., Walport, M. & Janeway, C. Janeway's immunobiology. 8th edn, (Garland Science, 2012).
2Perdue, S. & Humphrey, J. H. Immune System <https://www.britannica.com/science/immune-system/Classes-of-immunoglobulins#/media/1/283636/17661> (2020).
3Murray, P. R., Rosenthal, K. S. & Pfaller, M. A. Medical microbiology. 7th edn, (Elsevier Saunders, 2013).
4Chen, Y., Liu, Q. & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of medical virology 92, 418-423, doi:10.1002/jmv.25681.
5Mathur, G. & Mathur, S. (2020) Antibody Testing For Covid-19. Am J Clin Pathol, doi:10.1093/ajcp/aqaa082.
6FDA, U. S. (2020) FAQs on diagnostic testing for SARS CoV-2, <https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-diagnostic-testing-sars-cov-2>.
7Sood, N. et al. (2020) Seroprevalence of SARS-CoV-2-Specific Antibodies Among Adults in Los Angeles County, California, on April 10-11, 2020. JAMA, doi:10.1001/jama.2020.8279.
8Tong, P. B., Lin, L. Y. & Tran, T. H. (2020) Coronaviruses pandemics: Can neutralizing antibodies help? Life Sci, 117836, doi:10.1016/j.lfs.2020.117836.
9FDA, U. S. (2020) EUA Authorized Serology Test Performance, <https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance>.
1010FDA, U. S. (2020) Emergency Use Authorizations, <https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations>.
11Cai, X. F. et al. (2020) A Peptide-based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019 (COVID-19). J Infect Dis, doi:10.1093/infdis/jiaa243.
12Ni, L. et al. (2020) Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity, doi:10.1016/j.immuni.2020.04.023.
13Kontou, P. I., Braliou, G. G., Dimou, N. L., Nikolopoulos, G. & Bagos, P. G. (2020) Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis. Diagnostics (Basel) 10, doi:10.3390/diagnostics10050319.
14Lou, B. et al. (2020) Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J, doi:10.1183/13993003.00763-2020.
15Suhandynata, R. T. et al. (2020) Longitudinal Monitoring of SARS-CoV-2 IgM and IgG Seropositivity to Detect COVID-19. J Appl Lab Med, doi:10.1093/jalm/jfaa079.
16Liu, L. et al. (2020) A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes and infection, doi:10.1016/j.micinf.2020.05.008.
17French, M. A. & Moodley, Y. (2020) The role of SARS-CoV-2 antibodies in COVID-19: Healing in most, harm at times. Respirology, doi:10.1111/resp.13852.
18Long, Q.-X. et al. (2020) Antibody responses to SARS-CoV-2 in patients with COVID-19. Nature Medicine, doi:10.1038/s41591-020-0897-1.
19Ma, H. et al. (2020) Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol, doi:10.1038/s41423-020-0474-z.
20Amanat, F. et al. (2020) A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med, doi:10.1038/s41591-020-0913-5 .
21Perera, R. A. et al. (2020) Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill 25, doi:10.2807/1560-7917.Es.2020.25.16.2000421.
22Stadlbauer, D. et al. (2020) SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr Protoc Microbiol 57, e100, doi:10.1002/cpmc.100.
23Zhao, J. et al. (2020) Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases, doi:10.1093/cid/ciaa344.
24Qu, J. et al. (2020) Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis, doi:10.1093/cid/ciaa489.
25Wölfel, R. et al. (2020) Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465-469, doi:10.1038/s41586-020-2196-x.
26Tré-Hardy, M. et al. (2020) Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin Chem Lab Med, doi:10.1515/cclm-2020-0594.
27Li, Z. et al. (2020) Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. Journal of medical virology, doi:10.1002/jmv.25727.
28Burbelo, P. D. et al. (2020) Detection of Nucleocapsid Antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. J Infect Dis, doi:10.1093/infdis/jiaa273.
29Walls, A. C. et al. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, doi:10.1016/j.cell.2020.02.058.
30Shi, R. et al. (2020) A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature, doi:10.1038/s41586-020-2381-y.
31Ju, B. et al. (2020) Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature, doi:10.1038/s41586-020-2380-z.
32Premkunar, L. et al. (2020) The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5, doi:10.1126/sciimmunol.abc8413.
33Rogers, T.F. et al. (2020) Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. doi:10.1126/science.abc7520.
34Seydoux, E. et al. (2020) Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. doi:10.1016/j.immuni.2020.06.001.
35Brouwer, P. J. M. et al. (2020) Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. doi:10.1126/science.abc5902.
36Nile, S. H. et al. (2020) COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev, doi:10.1016/j.cytogfr.2020.05.002.
37Lee, Cheryle Yi-Pin et al. (2020) Serological Approaches for COVID-19 : Epidemiologic Perspsective on Surveillance and Control. Frontiers in Immunology 11, 879. doi:10.3389/fimmu.2020.00879
Coronavirus Products and Support
Information on testing for COVID-19 infection and antibodies is available on our SARS-CoV-2 Serology and PCR Testing page.