Methods of Detection
There are several methods that can be used to detect microsatellite instability (MSI) or deficient DNA mismatch repair (dMMR). The two most common methods are polymerase chain reaction (PCR) and immunohistochemistry (IHC). MSI testing by PCR is the accepted gold standard method for MSI detection(1). Learn more about the various methods and how they compare.
Comparison of MSI by PCR and MMR by IHC
MSI by PCR
Mononucleotide repeat microsatellite sequences are particularly sensitive to transcription errors, making them ideal targets for measurement by PCR amplification. To detect MSI, fluorescently labeled primers are used to amplify fragments from the tumor and matched normal samples. The amplified fragments are separated by size using capillary electrophoresis, and the fluorescent labelling allows them to be separated by the color of the fluorescent tag. Multiplexing the fluorescent labels allows many different fragments of similar size to be detected in the same reaction. Changes in sizing indicate that there is microsatellite instability, and tumors that contain this microsatellite instability are referred to MSI-high or MSI-H.
Key points about MSI by PCR
- Directly measures changes in DNA caused by loss of MMR protein
- Functional measure of mismatch repair deficiency that detects loss in MMR repair function, regardless of cause
- Does not identify the MMR gene to investigate
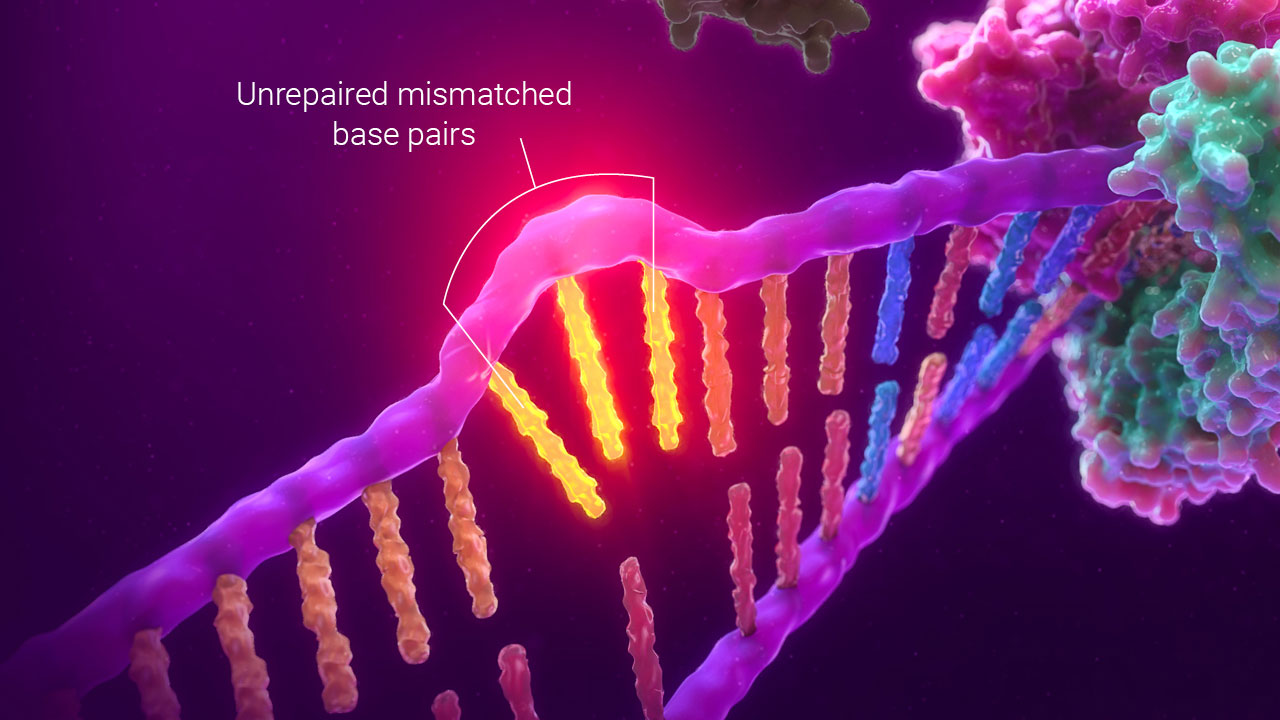
MSI by PCR uses changes in fragment size caused by unrepaired mismatched bases to detect a defect in mismatch repair function.
MMR by IHC
The presence or absence of MMR proteins can be detected using IHC, which is a histological technique using labeled, protein-specific antibodies to detect proteins in tissue samples. Each protein is detected using a separate tissue sample. In the case of the major mismatch repair proteins, tumors that have loss of one or more of the mismatch repair proteins are referred to as dMMR, because deficiency in mismatch repair is implied.
MMR protein expression is not necessarily a conclusive measure of MMR function. There can be a loss in the function of these proteins without a corresponding loss of the protein in the cell, with 5–10% of proteins retaining antigenicity when they are not functional(2).
Key points about MMR by IHC
- Detects the presence or absence of MMR proteins
- Not a conclusive measure of MMR function
- Shows which MMR gene to investigate
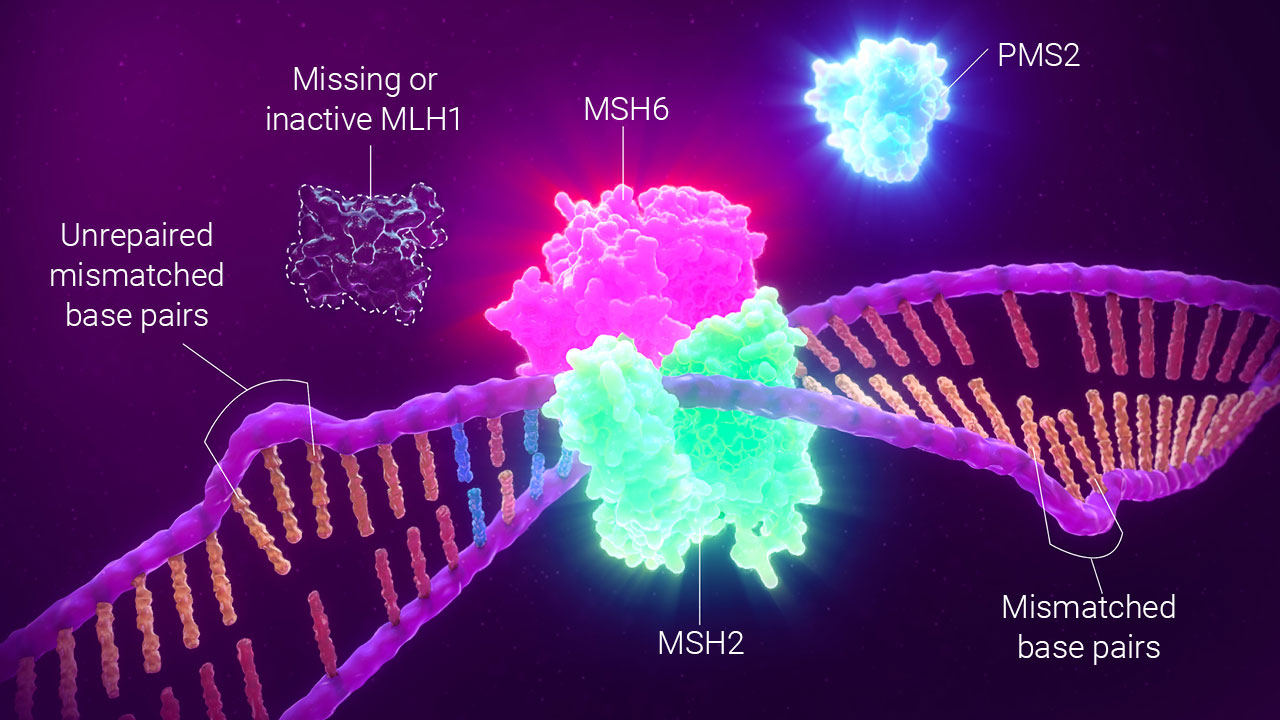
dMMR by IHC detects the presence or absence of MMR proteins.
MSI Analysis by Next Generation Sequencing (NGS)
MSI analysis by NGS determines sequences of microsatellites and then compares them to either matched normal tissue or a consensus sequence. Differences in the microsatellite sequence length indicate instability. NGS uses algorithms to determine if the amount of instability detected is significant and then assigns MSI-H or MSS (microsatellite stable) status. In some cases, computational challenges inherent to NGS analysis of microsatellite homopolymers may mean that no call can be made. There is currently no consensus on which loci or what data analysis pipeline and cutoffs should be used for MSI determination by NGS so there are many different approaches used in the field.
Key points about NGS
- Can be automated
- Requires highly stringent DNA quality
- Lacks standard, validated parameters (sequencing technology, MSI algorithm, Gene Panel, etc.) for detecting dMMR in tumors
PCR and IHC Co-Testing for MSI/dMMR
MSI by PCR and dMMR by IHC offer fundamentally different information about tumor samples(3). Specifically, IHC provides information about the MMR proteins expressed in the sample, while MSI by PCR measures MMR function by detecting changes in DNA that results when major MMR function is lost.
When used in parallel, PCR and IHC testing methods can increase the overall number of correctly characterized tumors to over 99%.

Clinical Co-Testing Guidelines
“Although loss of MMR protein immunoreactivity is generally detected in dMMR colorectal cancer, normal immunoreactivity can be seen in up to 10% of dMMR cases; therefore, MSI DNA testing may be performed either stepwise or as a concurrent test”(6).
Comparison of PCR, IHC and NGS Detection Methods
| Overview | Detects the functional failure of tumor MMR proteins, resulting in changes (instability) in microsatellite allele length | Detects the presence or absence of tumor MMR proteins using monoclonal antibodies against the major mismatch repair genes | Detects mutations within microsatellite sequences of tumor samples |
| Cost | $45/sample | $50–70/slide (x 4 slides) | $1–3K/sample (variable) |
| Throughput | Up to 96 samples (med to high) | Low to medium | Variable (more information = lower throughput) |
| Sample Input | 1ng (<1 FFPE curl) | 4 slides | 10–20 slides |
| Turnaround Time | ~10 Hours | 1–3 days | 2–3 weeks (2–6 weeks if send-out) |
| False Negative Rate | 0.3–4%(5) | 5–10%(6) | Variable (not standardized) |
| Multiplexed | Yes | No (must do separate slides) | Yes |
| Markers Analyzed | 5 mononucleotide loci (NCI recommended, Revised Bethesda Panel)(8) | 4–5 MMR proteins (MSH6, MSH2, PMS2, MLH1) | No standard panel or analysis method established or recommended |
| Pros | Functional test for dMMR, low false negative rate, proven technology, NCI recommended standardized loci panel available , 225+ peer-reviewed publications, minimal sample needed | Shows which genes to investigate, familiar technology | Provides more information besides just MMR (MSI), high throughput |
| Limitations | Only characterizes MSI, requires molecular training, does not indicate MMR genes to investigate | 5–10% false negative rate, uses more sample(6), throughput, indirect measure of dMMR | Large sample requirements (>20ng DNA), lack of standardization, requires advanced molecular training, technical issues lead to misclassification or false results, highly stringent DNA quality requirements |
References:
- Li, K. et al. (2020) Microsatellite instability: a review of what the oncologist should know. Cancer Cell International. 20, 16.
- Funkhouser, W.K. Jr. et al. (2012) Relevance, Pathogenesis, and Testing Algorithm for Mismatch Repair–Defective Colorectal Carcinomas: A Report of the Association for Molecular Pathology J. Mol. Diagnostics. 14(2), 91–103.
- Zhang, et al. (2008) Immunohistochemistry versus Microsatellite Instability Testing for Screening Colorectal Cancer Patients at Risk for Hereditary Nonpolyposis Colorectal Cancer Syndrome: Part II. The Utility of Microsatellite Instability Testing. J. Mol. Diagnostics. 10, 301–7.
- Dudley, J.C. et al. (2016) Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin. Cancer Res. 22, 813–820.
- Based on an internal analysis of publications comparing MSI-PCR v. IHC-dMMR in colorectal cancer from 2004–2018. Literature bundle available from Promega Medical Affairs upon request.
- Sepulveda, A.R. et al. (2017) Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diag. 19, 187–225.
- Luchini, C. et al. (2019) ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Annals of Oncol. 30, 1232–1243.
- Umar et al. (2004) Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 96, 261–8.