Quantifying Protein Abundance at Endogenous Levels
Promega Corporation
Publication Date: 9/2017
Abstract
Control of protein abundance is one of the master regulators of cellular physiology. Expression levels change in response to a variety of cellular cues and alterations in protein degradation rates change stability and protein abundance in the cell. Understanding normal changes in cellular protein abundance is key to understanding basic cellular biology and the dysregulation that can underlie many disease states. This article describes use of HiBiT protein tagging system to precisely quantify cellular protein abundance. Results illustrate detection of stabilization of HIF1A and targeted degradation of BRD4. The HiBiT tag was introduced into the cell using a CRISPR/Cas9 method, allowing tagging of proteins expressed at endogenous levels, minimizing artifacts and providing a simple method to detect and quantify any tagged protein.
Common approaches for quantifying cellular protein abundance include mass spectrometry analysis or antibody-based approaches such as ELISA and western blotting. Epitope tagging methods, widely used for recombinant protein studies, are appealing because the small tags can easily be fused to a protein of interest and detected with specific, high-quality antibodies. However, these methods are limited in throughput or sensitivity, require high-quality antibodies and may only yield semi-quantitative results. Promega recently developed an alternative to classic epitope tagging approaches that brings the simplicity and sensitivity of bioluminescence to studies on protein abundance.
Bioluminescent Tagging System Derived from Small and Sensitive NanoLuc® Technology
We have used NanoLuc® Binary Technology (NanoBiT), a two-part complementation system based on NanoLuc® luciferase (1) , to develop a novel peptide tag that enables sensitive bioluminescent quantification of proteins with no antibody requirement. The tag, designated High BiT (HiBiT), is only 11 amino acids in length. HiBiT-tagged proteins are measured using detection reagents containing the peptide’s complementing polypeptide, Large BiT (LgBiT), which binds with high affinity to HiBiT (Kd ~1nM) reconstituting bright, luminescent enzyme activity. In contrast to antibody-based detection methods, the HiBiT tagging system provides a highly quantitative measure of protein abundance using simple add-mix-measure detection with over seven logs of linear dynamic range, making it compatible with both bench-level and high-throughput studies of protein abundance.
Measuring Endogenous Protein Expression
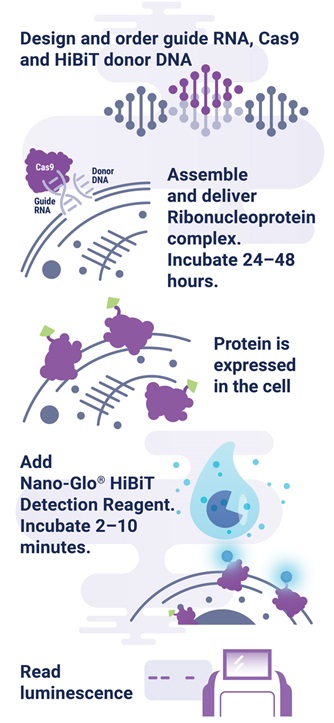
The HiBiT tagging system has the sensitivity to quantitatively measure proteins down to endogenous levels, even for proteins typically maintained at low levels in the cell. In addition, the small 11aa tag enables efficient integration of HiBiT into endogenous loci using a rapid CRISPR/Cas9-mediated genome editing strategy (Figure 1). The endogenous tagging system described here enables integration of the HiBiT tag at a precise genomic location using a method that parallels traditional vector-based transient transfection without the cloning requirement (2) . When used together, HiBiT and CRISPR/Cas9 allow for quantitative studies of protein abundance under endogenous expression conditions with a simple, quantitative detection workflow.
Figure 1. Endogenous HiBiT protein tagging system protocol. The 11aa tag enables efficient integration of HiBiT into endogenous loci using a rapid, cloning-free CRISPR/Cas9 gene-editing strategy as outlined. One can design and order guide RNA, Cas9, and HiBiT donor DNA from supplier such as IDT, assemble RNP complex, deliver RNP/donor to the cell, and detect genetic edit by luminescence.
HiBiT Can Quantify Protein Stabilization
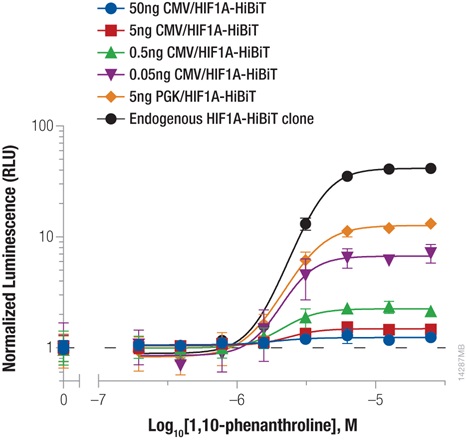
Hypoxia-inducible factor-1A (HIF1A) is a transcription factor involved in the response to hypoxic conditions that is thought to play an important role in cancer metabolism. Protein levels are kept low under normal basal conditions by prolyl hydroxylation and VHL-directed ubiquitination. Hypoxic conditions or chemical inhibition of prolyl hydroxylases with compounds such as 1,10-phenanthroline lead to accumulation of HIF1A protein for a rapid cellular response. The accumulation of HIF1A was measured by expressing HIF1A-HiBiT fusion from either CMV- or PGK-driven expression constructs using transient transfection or by insertion of HiBiT into the endogenous HIF1A loci using CRISPR/Cas9 gene editing (Figure 2). The low background and wide dynamic range of the HiBiT assay enable detection of the even the low, basal levels of endogenously expressed HIF1A.
We obtained the expected dose response increase in HIF1A abundance following increased treatment with 1,10-phenanthroline. We also observed that lower levels of protein expression led to a greater fold response of accumulation, with both expression from the weak PGK promoter and endogenous expression following CRISPR/Cas9 insertion displaying the largest fold change.
Figure 2: Stabilizing Hif1a-HiBiT by incubation with the hypoxia mimetic 1,10-phenanthroline. HeLa cells were transiently transfected with different amounts of CMV- or PGK-driven expression constructs for Hif1a-HiBiT, diluted in carrier DNA. In parallel, HiBiT was tagged to the endogenous locus in HeLa cells using CRISPR/Cas9, and a clone was isolated. Cells were plated in 96-well plates and treated the following day for 4 hours with a titration of 1,10-phenanthroline. The Nano-Glo® HiBiT Lytic Reagent was added to all wells, and luminescence was measured after 10 minutes as in Figure 1. Data were normalized to untreated cells.
HiBiT Can Quantify Targeted Protein Degradation
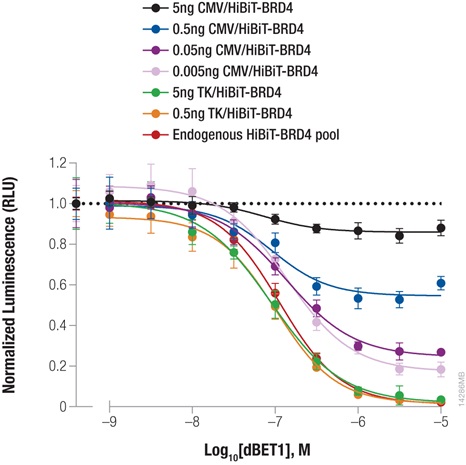
Bromodomain-containing protein, BRD4, is one attractive target for cancer therapeutics. Compound dBET1 links BRD4 to cereblon, a component of the E3 ligase complex, to induce its degradation. The function of dBET1 in targeting BRD4 for degradation was initially demonstrated using immunoblotting with a primary antibody that recognized the BRD4 protein (3) . We have used HiBiT tagging to quantify the degradation of BRD4 following dBET1 treatment by expressing HiBiT-tagged BRD4 in HEK293 cells, either by transiently transfecting different dilutions of TK or CMV expression constructs or by introducing the HiBiT tag at the endogenous locus using CRISPR/Cas9 editing (Figure 3). Following a 2-hour incubation with dBET1, BRD4 abundance was significantly reduced in the cells. Interestingly, the CRISPR/Cas9-derived cell pool expressing HiBiT-BRD4 at endogenous levels displayed significantly greater degradation compared to transiently expressed protein, demonstrating how overexpression artifacts can hinder studies of protein degradation. The sensitivity of the HiBiT system enabled us to perform these experiments in a pool of edited cells, as opposed to further generating a clonal population.
Figure 3: Targeted degradation of HiBiT-BRD4 by incubating with compound dBET1. HEK293 cells were transiently transfected with different amounts of HiBiT-BRD4 expression constructs with either the strong CMV promoter or the weak TK promoter, diluted in carrier DNA to keep the DNA amount constant. In parallel, a pool of HEK293 cells expressing HiBiT-BRD4 from the endogenous locus were generated using CRISPR/Cas9. Cells were plated in 96-well plates and treated the following day for 2 hours with a titration of dBET1 to induce degradation. The Nano-Glo Lytic Reagent was added to all wells, and luminescence was measured after 10 minutes as in Figure 2. Background was subtracted and luminescence data were normalized to untreated cells.
Summary
The HiBiT tagging system provides a simple, bioluminescent method to precisely quantify cellular protein abundance. For both stabilization of HIF1A and targeted degradation of BRD4, we observed the importance of maintaining low expression levels to achieve the most robust assay response. This observation highlights the importance of low expression levels in limiting experimental artifacts and maintaining proper stoichiometry with interacting proteins such as those involved in the degradation machinery. HiBiT-tagged proteins can be expressed either from transfected expression constructs diluted to appropriate levels or from the endogenous locus by knocking in with CRISPR/Cas9. Bright signal output and sensitivity of the HiBiT system enable quantitative detection even at these low levels using a simple detection format.
Article References
- Dixon, A.S. et al. (2016) NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–8.
- Schwinn, M. et al. (2018) CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem. Biol. 13, 467–74.
- Winter, G.E. et al. (2015) Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–81.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Landreman, A. and Eggers, C. Quantifying Protein Abundance at Endogenous Levels. [Internet] 9/2017. [cited: year, month, date]. Available from: https://worldwide.promega.com/es-es/resources/pubhub/2017/quantifying-protein-abundance-at-endogenous-levels/
American Medical Association, Manual of Style, 10th edition, 2007
Landreman, A. and Eggers, C. Quantifying Protein Abundance at Endogenous Levels. Promega Corporation Web site. https://worldwide.promega.com/es-es/resources/pubhub/2017/quantifying-protein-abundance-at-endogenous-levels/ Updated 9/2017. Accessed Month Day, Year.